Calcium »
PDB 8x8g-8yib »
8yf7 »
Calcium in PDB 8yf7: Cryo-Em Structure of Dragon Grouper Nervous Necrosis Virus-Like Particle at PH6.5 (2.82A)
Calcium Binding Sites:
The binding sites of Calcium atom in the Cryo-Em Structure of Dragon Grouper Nervous Necrosis Virus-Like Particle at PH6.5 (2.82A)
(pdb code 8yf7). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 3 binding sites of Calcium where determined in the Cryo-Em Structure of Dragon Grouper Nervous Necrosis Virus-Like Particle at PH6.5 (2.82A), PDB code: 8yf7:
Jump to Calcium binding site number: 1; 2; 3;
In total 3 binding sites of Calcium where determined in the Cryo-Em Structure of Dragon Grouper Nervous Necrosis Virus-Like Particle at PH6.5 (2.82A), PDB code: 8yf7:
Jump to Calcium binding site number: 1; 2; 3;
Calcium binding site 1 out of 3 in 8yf7
Go back to
Calcium binding site 1 out
of 3 in the Cryo-Em Structure of Dragon Grouper Nervous Necrosis Virus-Like Particle at PH6.5 (2.82A)
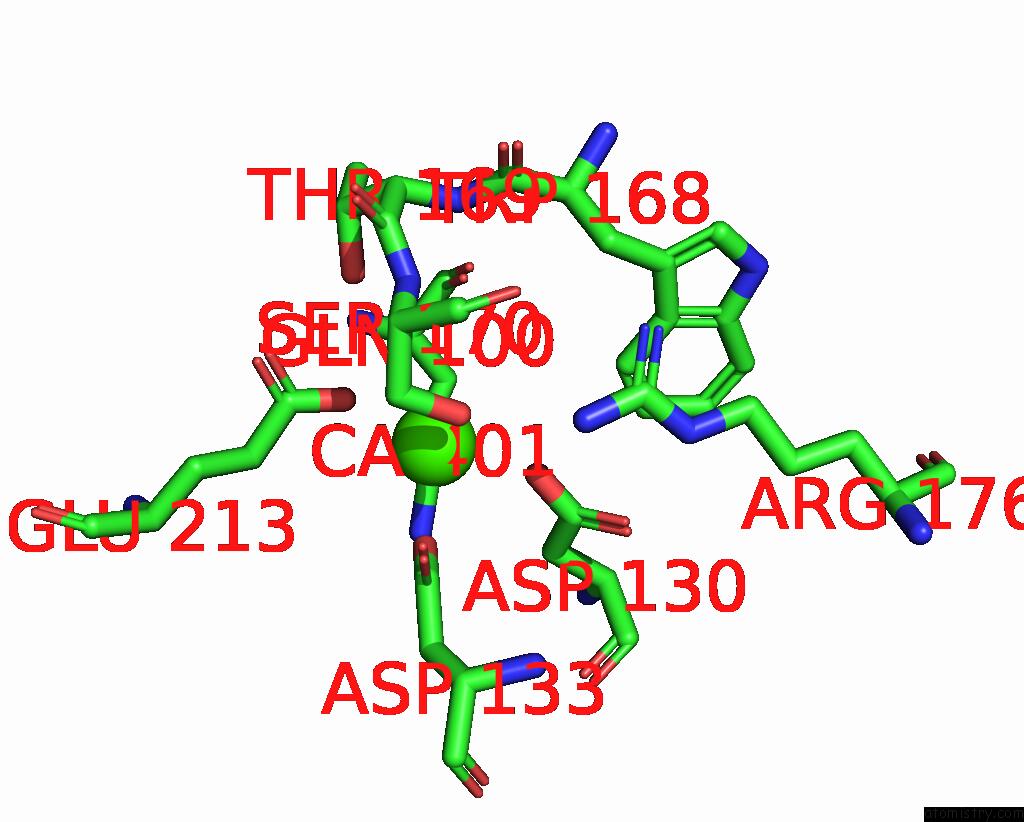
Mono view
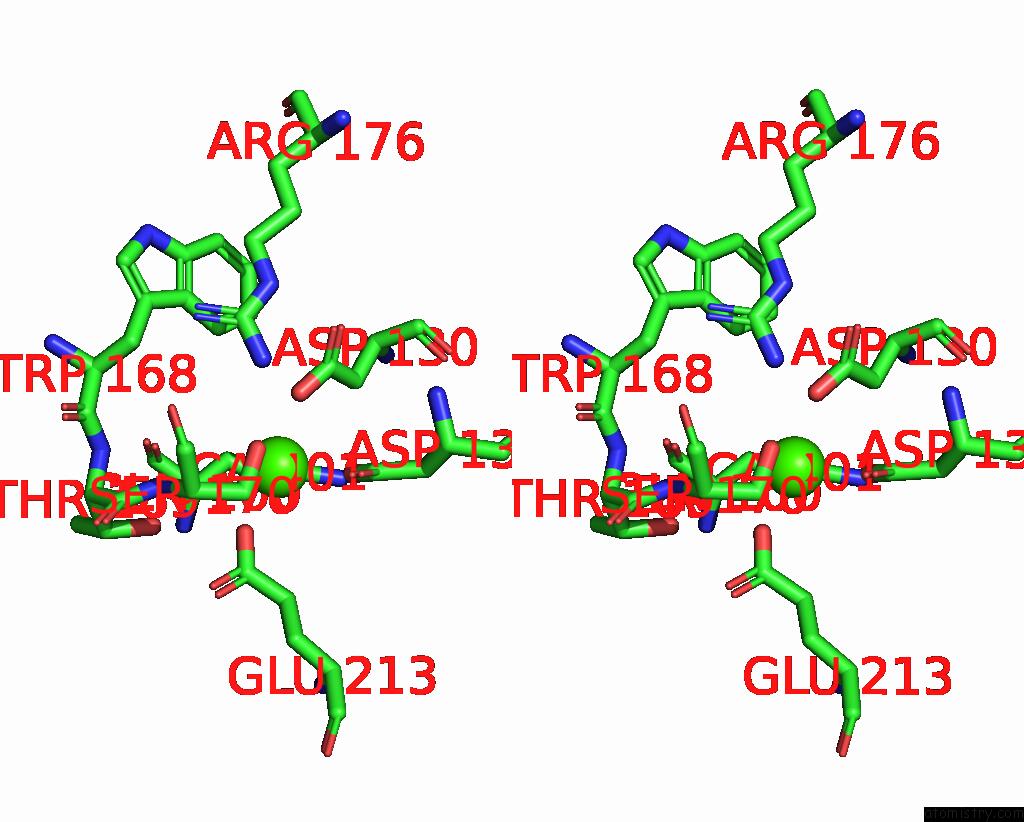
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Cryo-Em Structure of Dragon Grouper Nervous Necrosis Virus-Like Particle at PH6.5 (2.82A) within 5.0Å range:
|
Calcium binding site 2 out of 3 in 8yf7
Go back to
Calcium binding site 2 out
of 3 in the Cryo-Em Structure of Dragon Grouper Nervous Necrosis Virus-Like Particle at PH6.5 (2.82A)
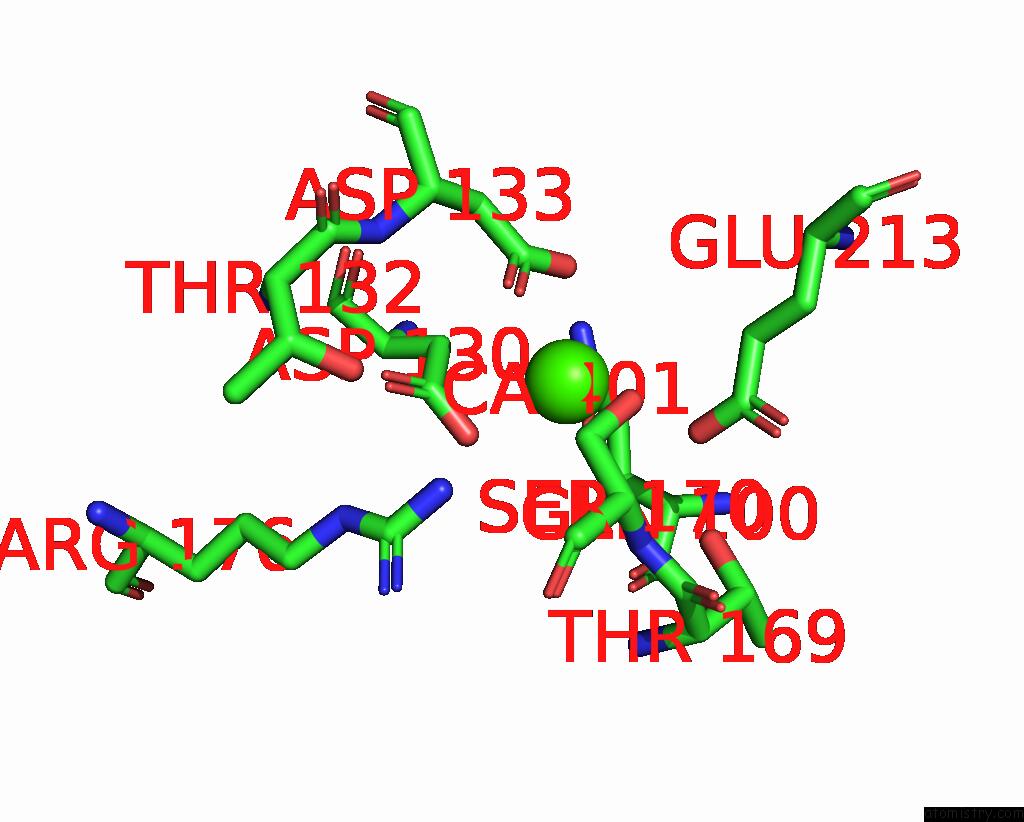
Mono view
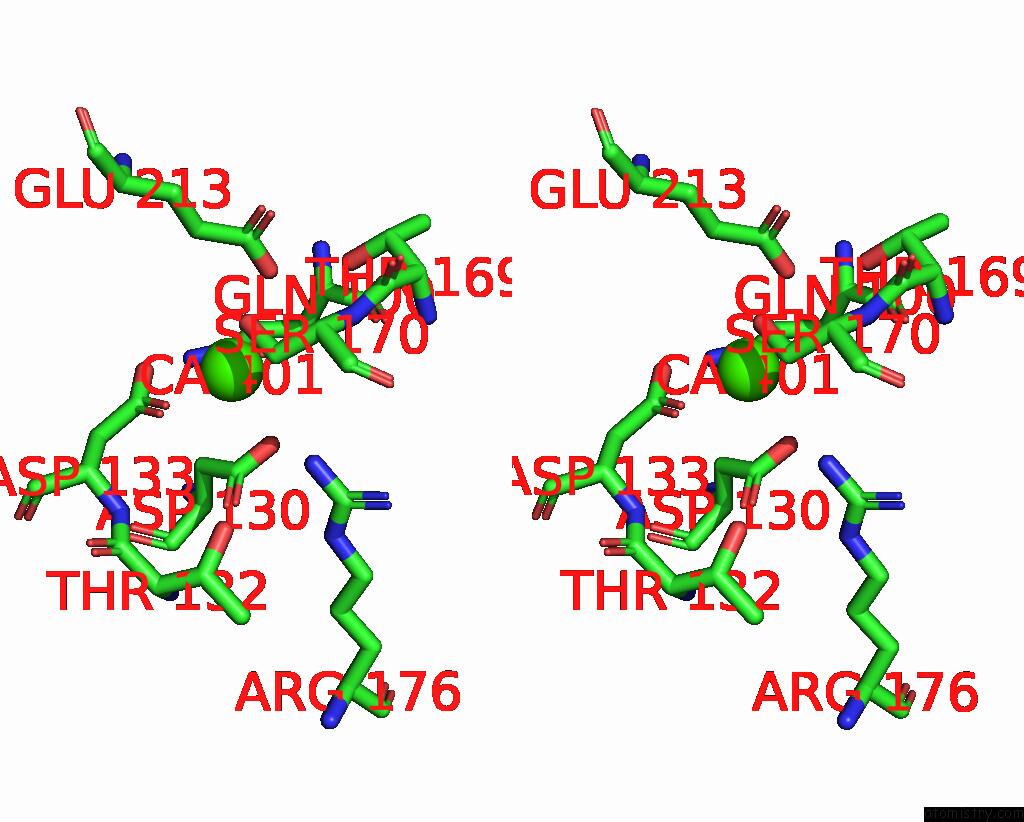
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Cryo-Em Structure of Dragon Grouper Nervous Necrosis Virus-Like Particle at PH6.5 (2.82A) within 5.0Å range:
|
Calcium binding site 3 out of 3 in 8yf7
Go back to
Calcium binding site 3 out
of 3 in the Cryo-Em Structure of Dragon Grouper Nervous Necrosis Virus-Like Particle at PH6.5 (2.82A)
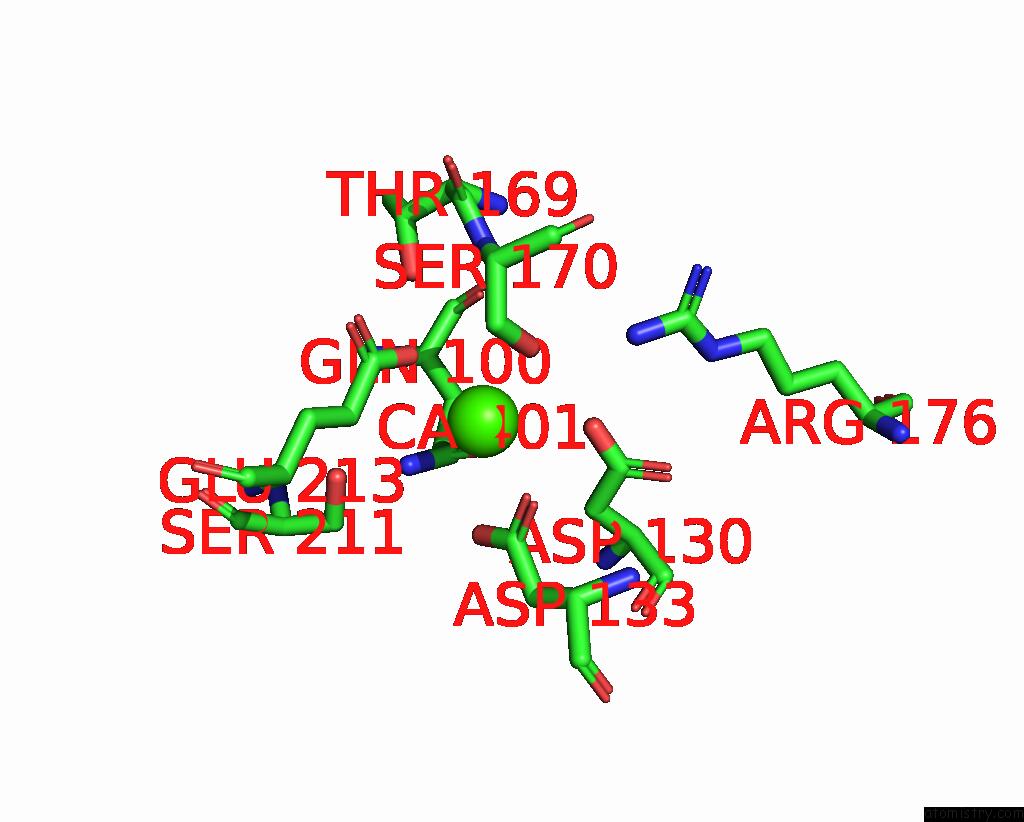
Mono view
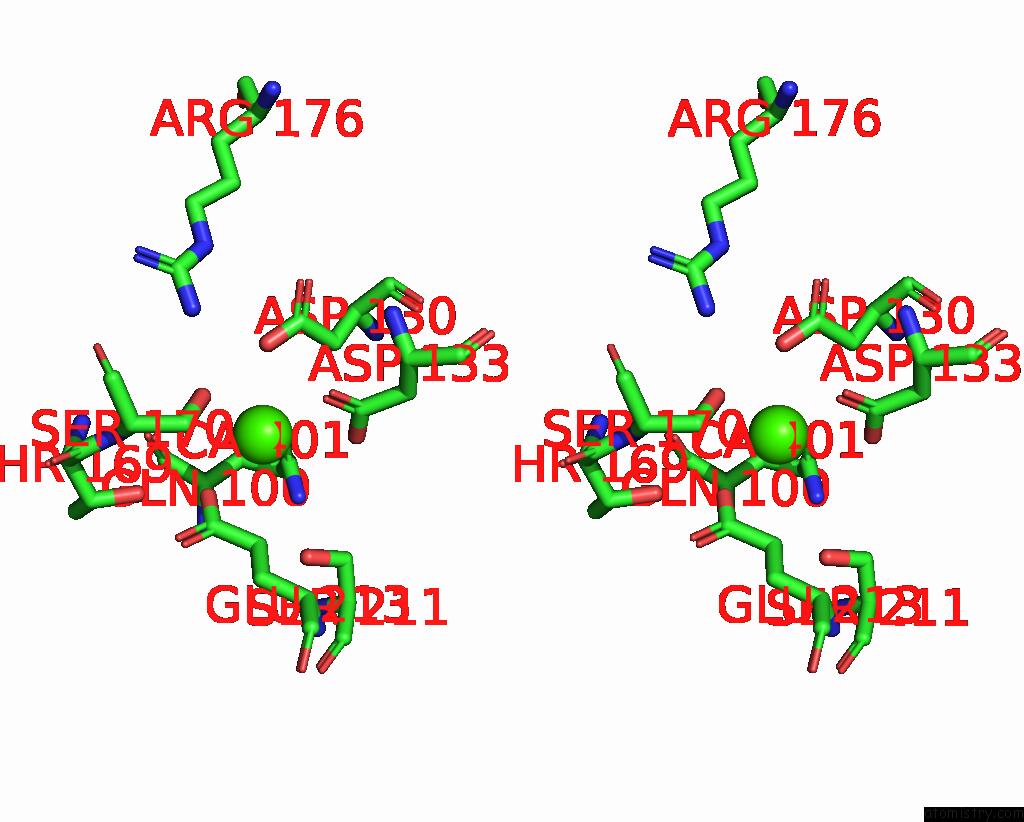
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 3 of Cryo-Em Structure of Dragon Grouper Nervous Necrosis Virus-Like Particle at PH6.5 (2.82A) within 5.0Å range:
|
Reference:
P.Sterbova,
C.H.Wang,
K.J.D.Carillo,
Y.C.Lou,
T.Kato,
K.Namba,
D.M.Tzou,
W.H.Chang.
Molecular Mechanism of pH-Induced Protrusion Configuration Switching in Piscine Betanodavirus Implies A Novel Antiviral Strategy. Acs Infect Dis. 2024.
ISSN: ESSN 2373-8227
PubMed: 39087906
DOI: 10.1021/ACSINFECDIS.4C00407
Page generated: Thu Jul 10 08:31:31 2025
ISSN: ESSN 2373-8227
PubMed: 39087906
DOI: 10.1021/ACSINFECDIS.4C00407
Last articles
F in 4E91F in 4E5D
F in 4E1V
F in 4DLE
F in 4E28
F in 4DZ9
F in 4E1N
F in 4DWV
F in 4DZ7
F in 4DXH