Calcium »
PDB 8x8g-8yib »
8yf8 »
Calcium in PDB 8yf8: Cryo-Em Structure of Dragon Grouper Nervous Necrosis Virus-Like Particle at PH5.0 (3.52A)
Calcium Binding Sites:
The binding sites of Calcium atom in the Cryo-Em Structure of Dragon Grouper Nervous Necrosis Virus-Like Particle at PH5.0 (3.52A)
(pdb code 8yf8). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 3 binding sites of Calcium where determined in the Cryo-Em Structure of Dragon Grouper Nervous Necrosis Virus-Like Particle at PH5.0 (3.52A), PDB code: 8yf8:
Jump to Calcium binding site number: 1; 2; 3;
In total 3 binding sites of Calcium where determined in the Cryo-Em Structure of Dragon Grouper Nervous Necrosis Virus-Like Particle at PH5.0 (3.52A), PDB code: 8yf8:
Jump to Calcium binding site number: 1; 2; 3;
Calcium binding site 1 out of 3 in 8yf8
Go back to
Calcium binding site 1 out
of 3 in the Cryo-Em Structure of Dragon Grouper Nervous Necrosis Virus-Like Particle at PH5.0 (3.52A)
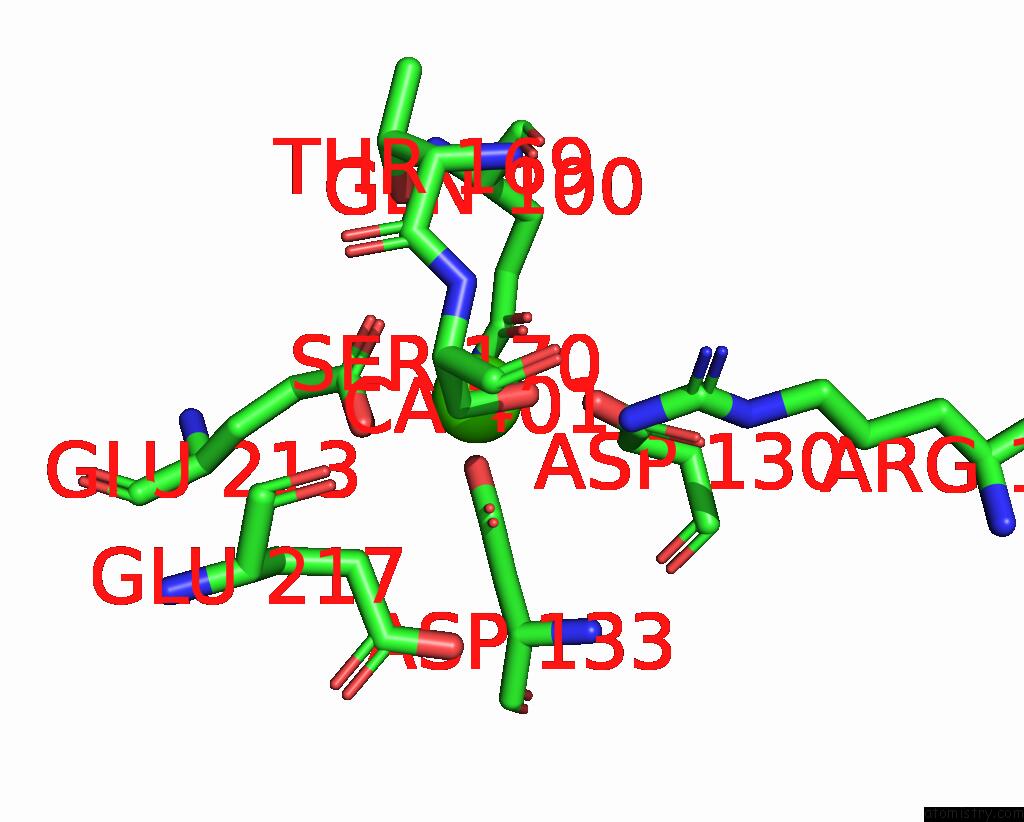
Mono view
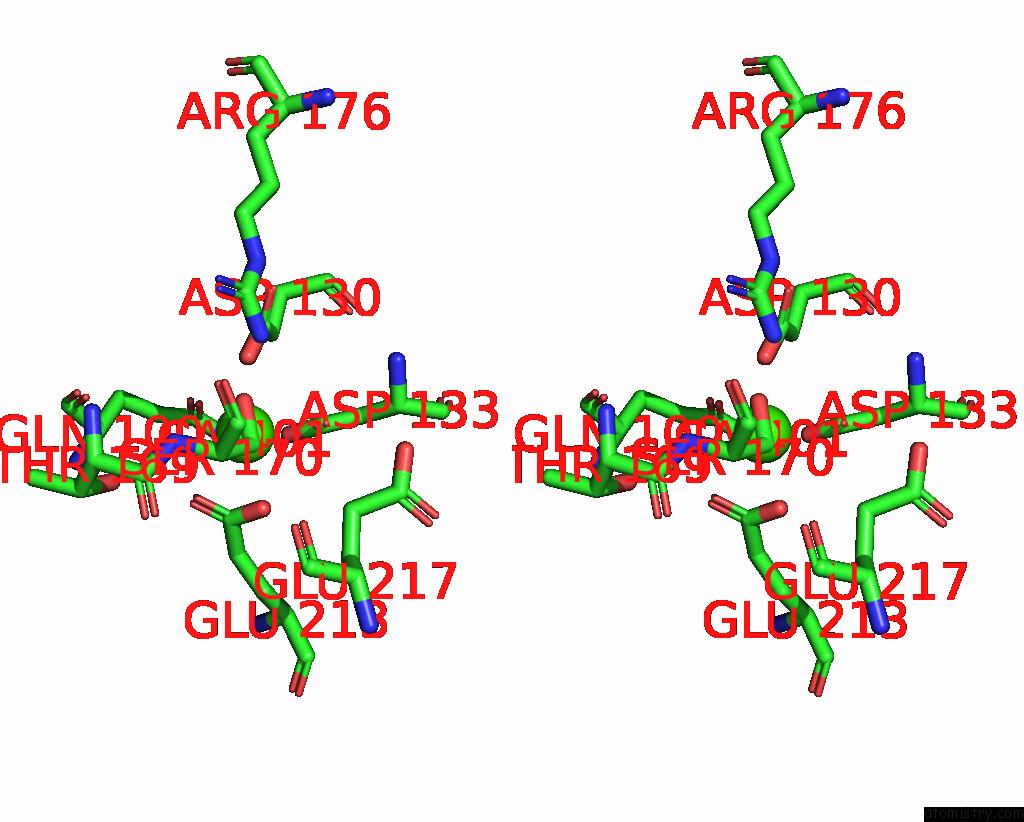
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Cryo-Em Structure of Dragon Grouper Nervous Necrosis Virus-Like Particle at PH5.0 (3.52A) within 5.0Å range:
|
Calcium binding site 2 out of 3 in 8yf8
Go back to
Calcium binding site 2 out
of 3 in the Cryo-Em Structure of Dragon Grouper Nervous Necrosis Virus-Like Particle at PH5.0 (3.52A)
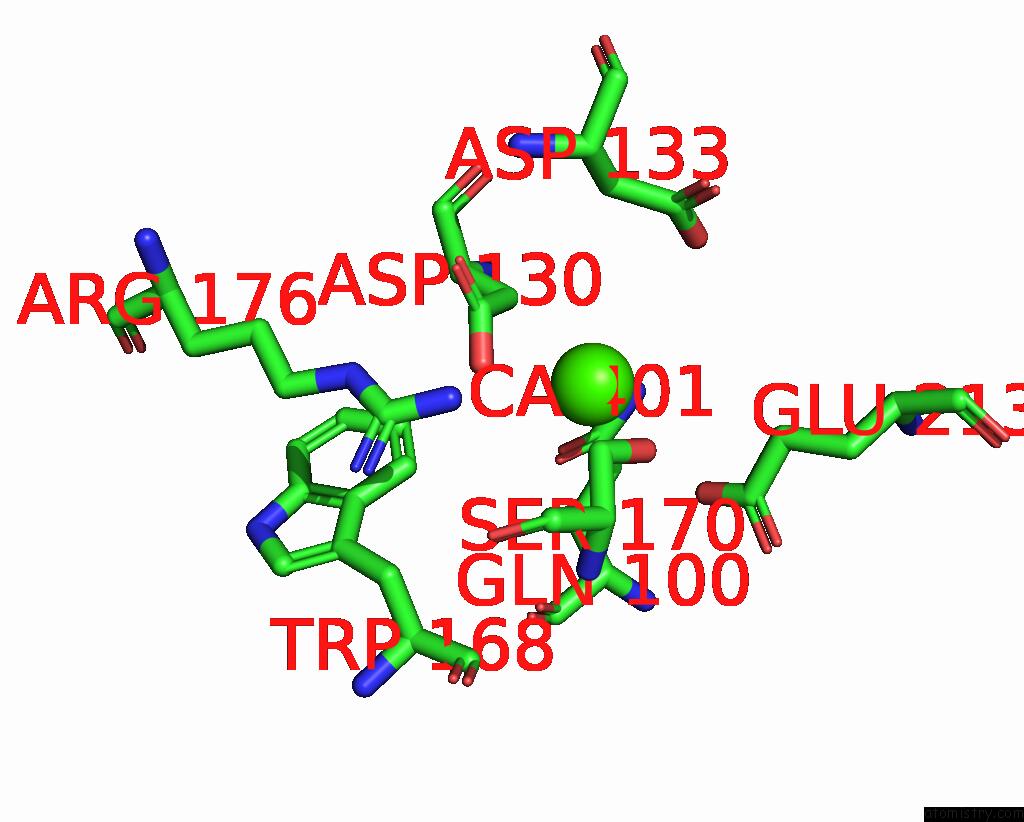
Mono view
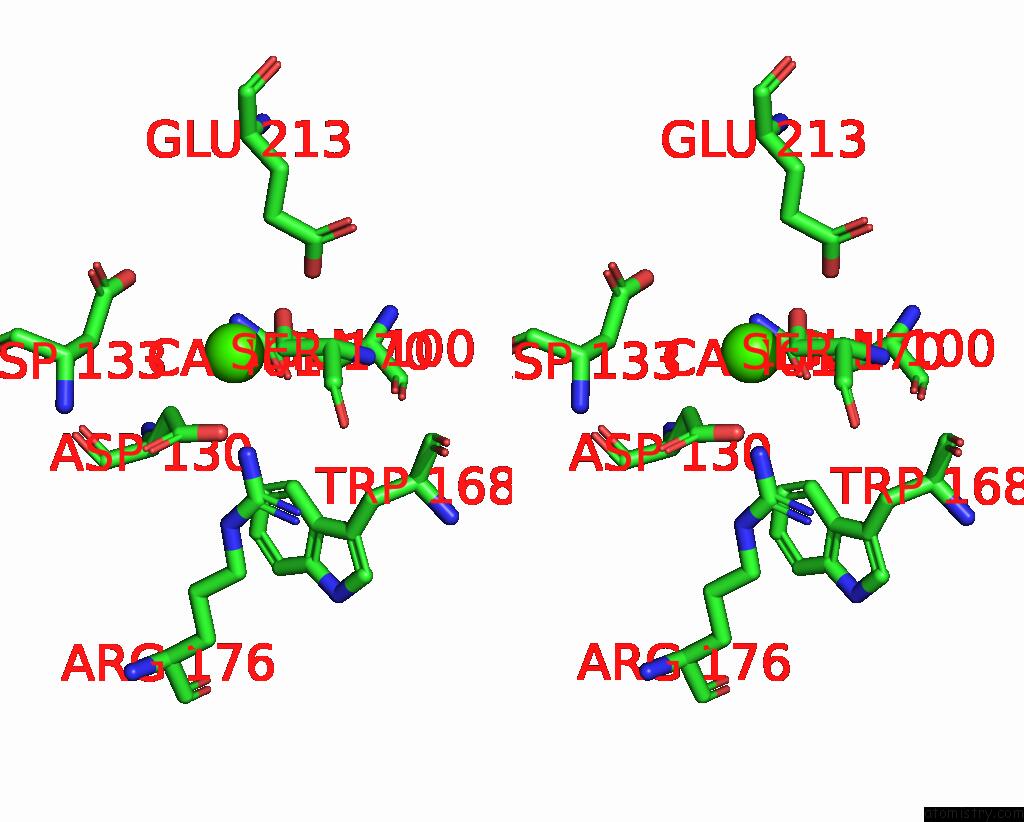
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Cryo-Em Structure of Dragon Grouper Nervous Necrosis Virus-Like Particle at PH5.0 (3.52A) within 5.0Å range:
|
Calcium binding site 3 out of 3 in 8yf8
Go back to
Calcium binding site 3 out
of 3 in the Cryo-Em Structure of Dragon Grouper Nervous Necrosis Virus-Like Particle at PH5.0 (3.52A)
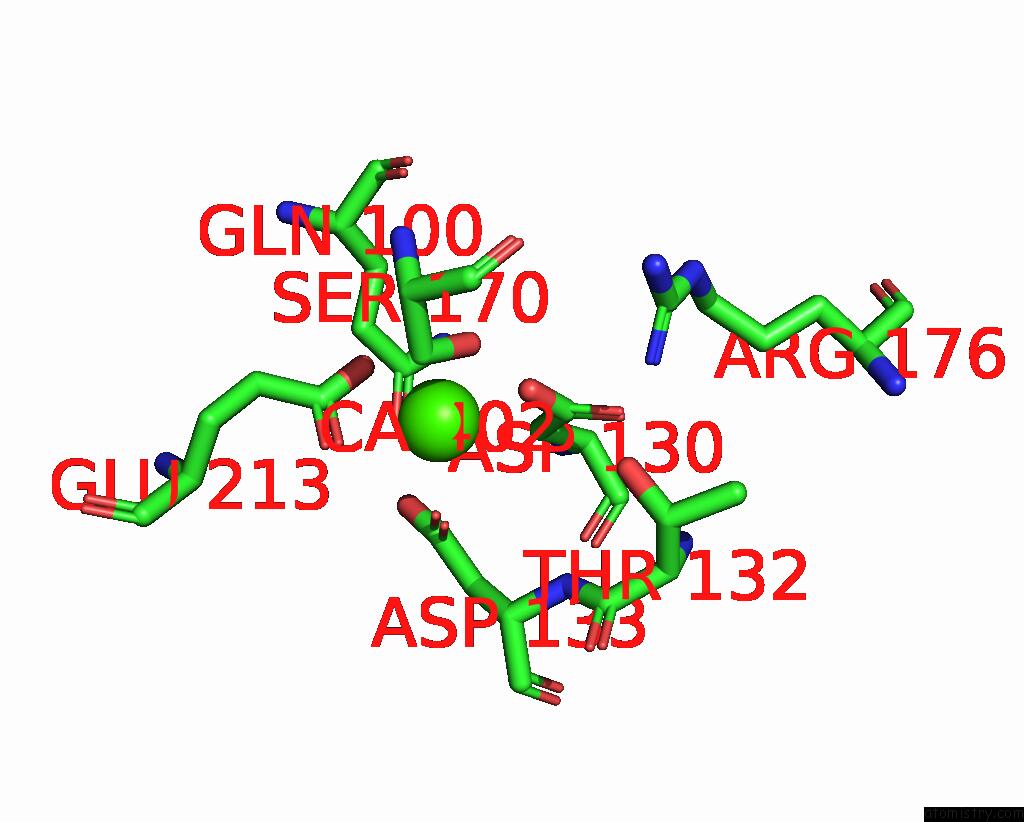
Mono view
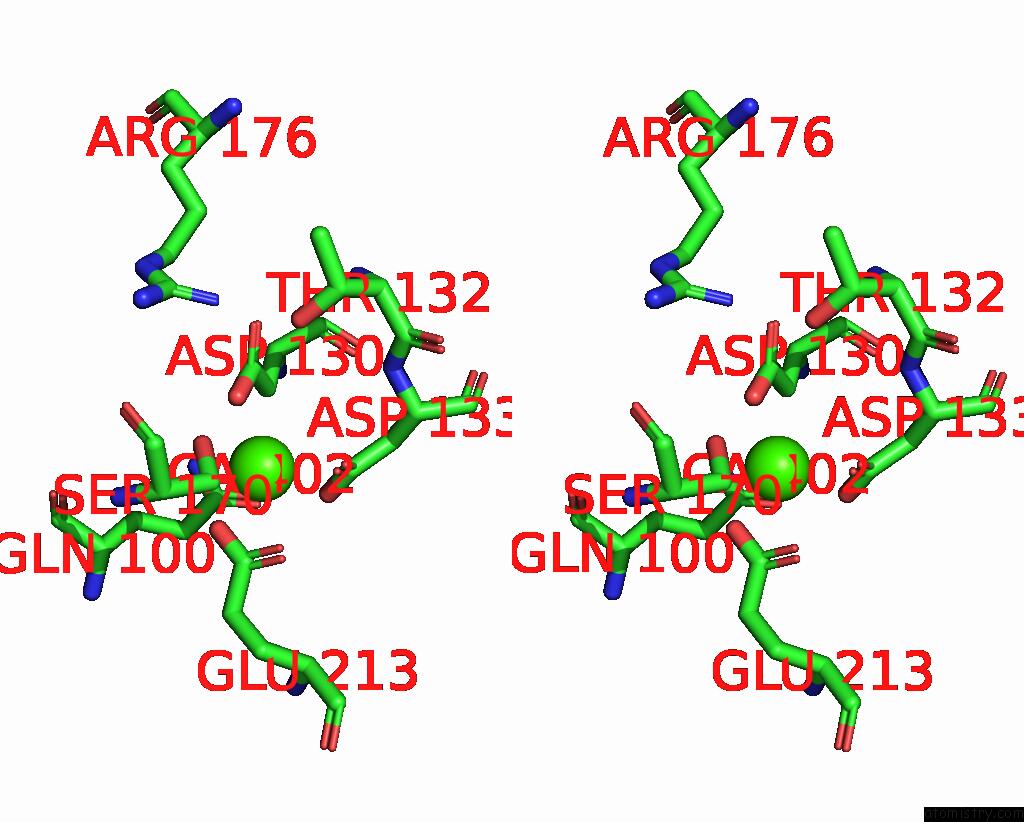
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 3 of Cryo-Em Structure of Dragon Grouper Nervous Necrosis Virus-Like Particle at PH5.0 (3.52A) within 5.0Å range:
|
Reference:
P.Sterbova,
C.H.Wang,
K.J.D.Carillo,
Y.C.Lou,
T.Kato,
K.Namba,
D.M.Tzou,
W.H.Chang.
Molecular Mechanism of pH-Induced Protrusion Configuration Switching in Piscine Betanodavirus Implies A Novel Antiviral Strategy. Acs Infect Dis. 2024.
ISSN: ESSN 2373-8227
PubMed: 39087906
DOI: 10.1021/ACSINFECDIS.4C00407
Page generated: Thu Jul 10 08:31:52 2025
ISSN: ESSN 2373-8227
PubMed: 39087906
DOI: 10.1021/ACSINFECDIS.4C00407
Last articles
Fe in 2YXOFe in 2YRS
Fe in 2YXC
Fe in 2YNM
Fe in 2YVJ
Fe in 2YP1
Fe in 2YU2
Fe in 2YU1
Fe in 2YQB
Fe in 2YOO