Calcium »
PDB 9cmo-9der »
9cyh »
Calcium in PDB 9cyh: Cryo-Em Structure of DA03E17 Fab in Complex with Influenza Virus Neuraminidase From B/Colorado/06/2017
Enzymatic activity of Cryo-Em Structure of DA03E17 Fab in Complex with Influenza Virus Neuraminidase From B/Colorado/06/2017
All present enzymatic activity of Cryo-Em Structure of DA03E17 Fab in Complex with Influenza Virus Neuraminidase From B/Colorado/06/2017:
3.2.1.18;
3.2.1.18;
Calcium Binding Sites:
The binding sites of Calcium atom in the Cryo-Em Structure of DA03E17 Fab in Complex with Influenza Virus Neuraminidase From B/Colorado/06/2017
(pdb code 9cyh). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 4 binding sites of Calcium where determined in the Cryo-Em Structure of DA03E17 Fab in Complex with Influenza Virus Neuraminidase From B/Colorado/06/2017, PDB code: 9cyh:
Jump to Calcium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Calcium where determined in the Cryo-Em Structure of DA03E17 Fab in Complex with Influenza Virus Neuraminidase From B/Colorado/06/2017, PDB code: 9cyh:
Jump to Calcium binding site number: 1; 2; 3; 4;
Calcium binding site 1 out of 4 in 9cyh
Go back to
Calcium binding site 1 out
of 4 in the Cryo-Em Structure of DA03E17 Fab in Complex with Influenza Virus Neuraminidase From B/Colorado/06/2017
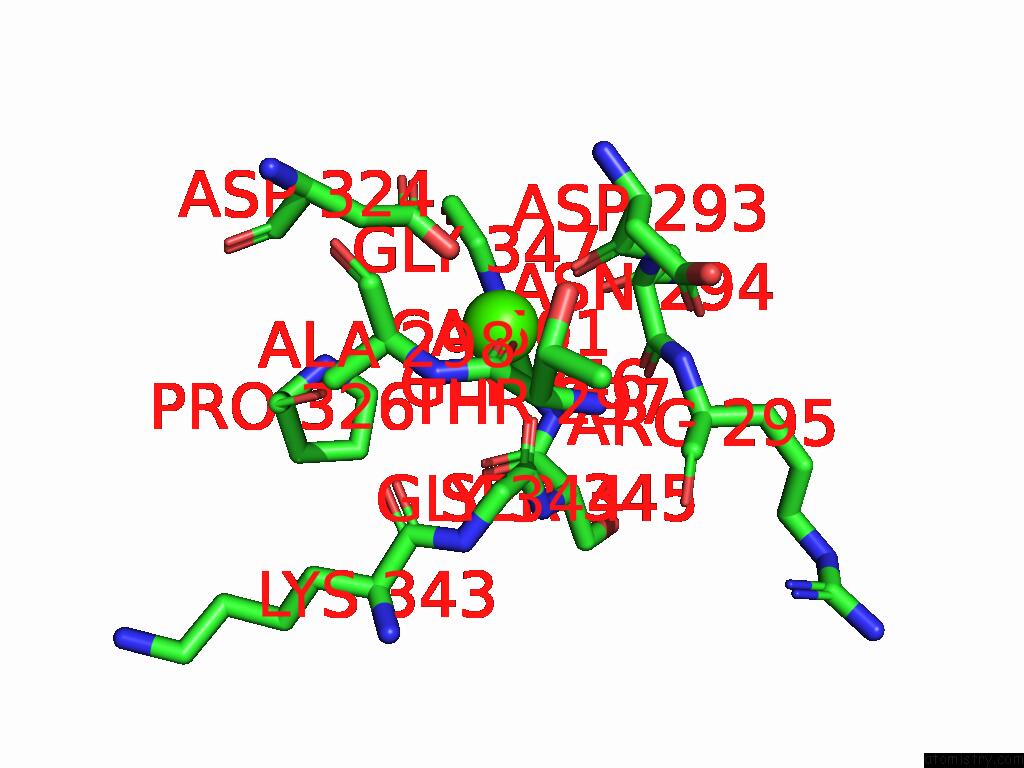
Mono view
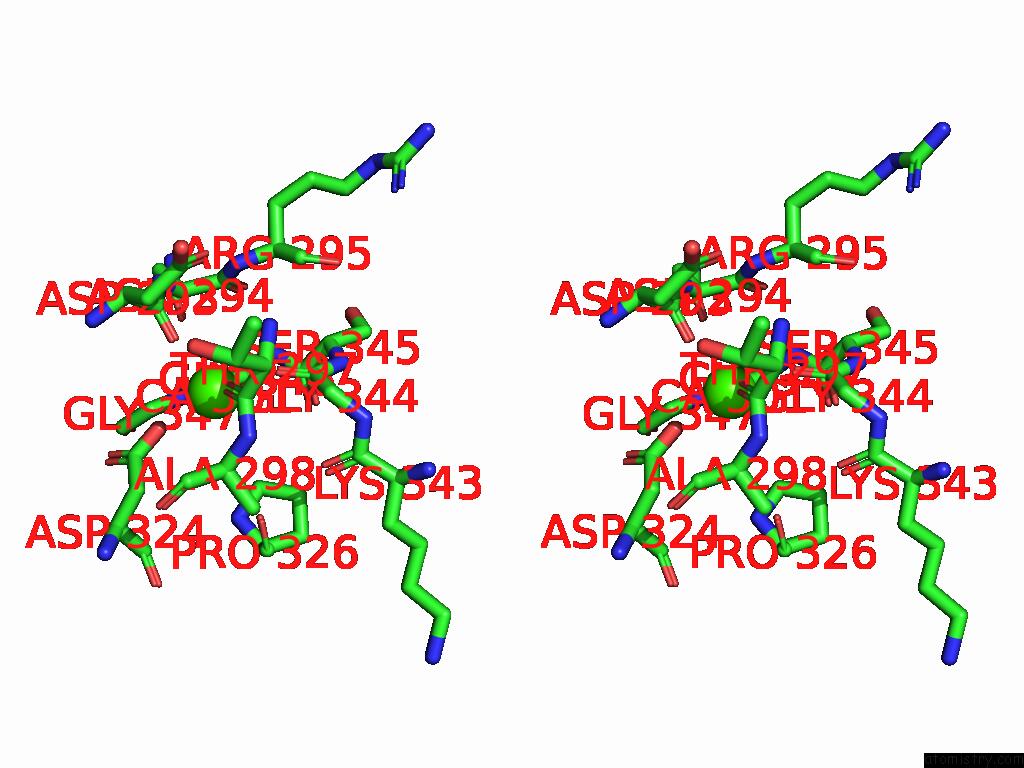
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Cryo-Em Structure of DA03E17 Fab in Complex with Influenza Virus Neuraminidase From B/Colorado/06/2017 within 5.0Å range:
|
Calcium binding site 2 out of 4 in 9cyh
Go back to
Calcium binding site 2 out
of 4 in the Cryo-Em Structure of DA03E17 Fab in Complex with Influenza Virus Neuraminidase From B/Colorado/06/2017
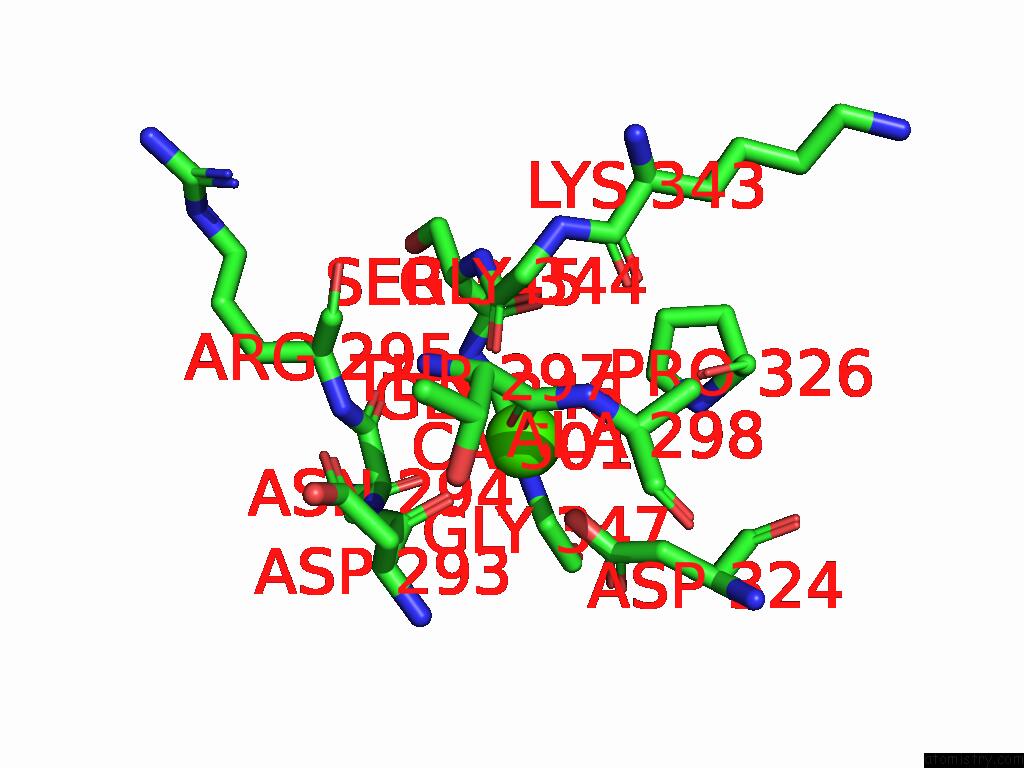
Mono view
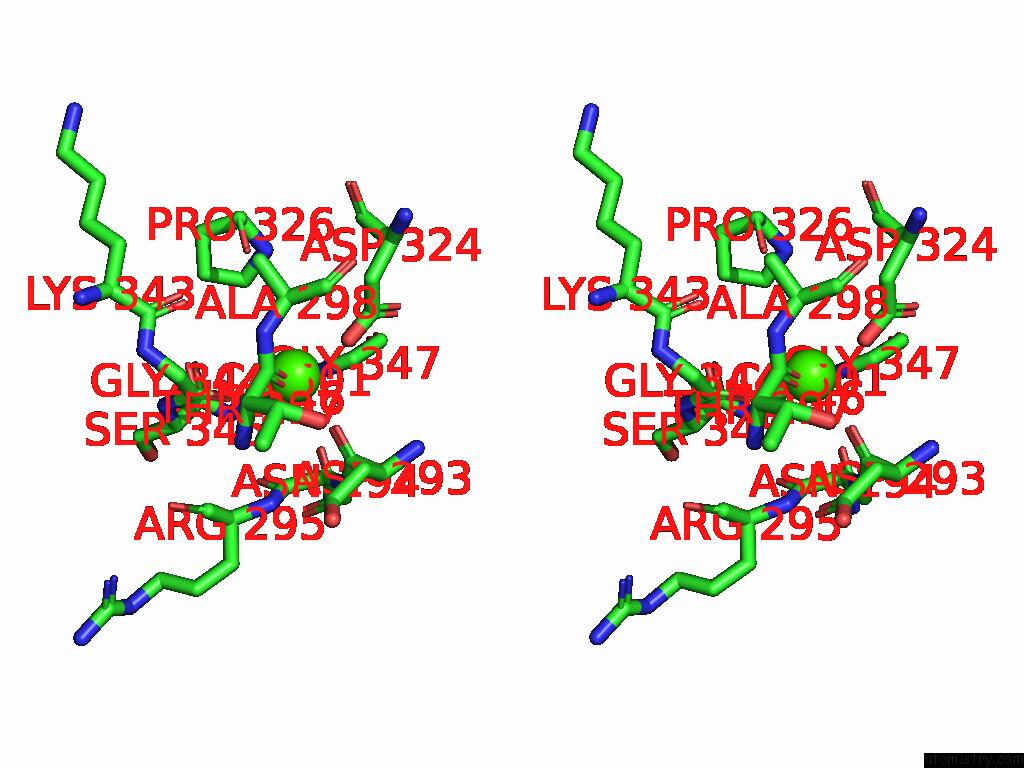
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Cryo-Em Structure of DA03E17 Fab in Complex with Influenza Virus Neuraminidase From B/Colorado/06/2017 within 5.0Å range:
|
Calcium binding site 3 out of 4 in 9cyh
Go back to
Calcium binding site 3 out
of 4 in the Cryo-Em Structure of DA03E17 Fab in Complex with Influenza Virus Neuraminidase From B/Colorado/06/2017
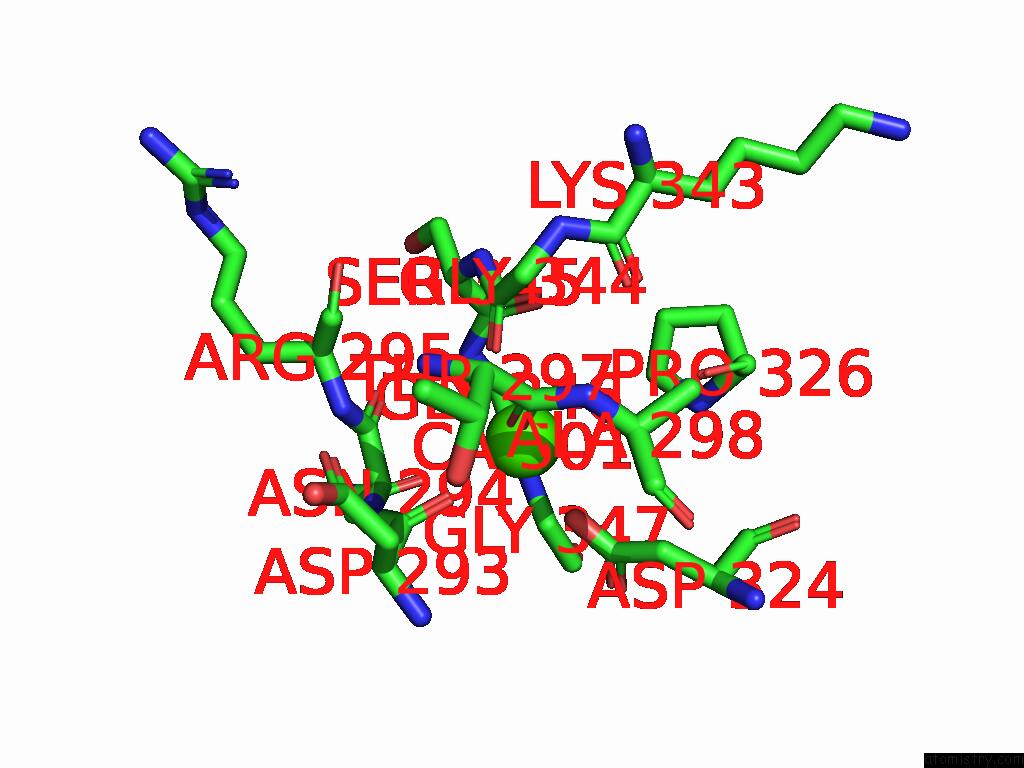
Mono view
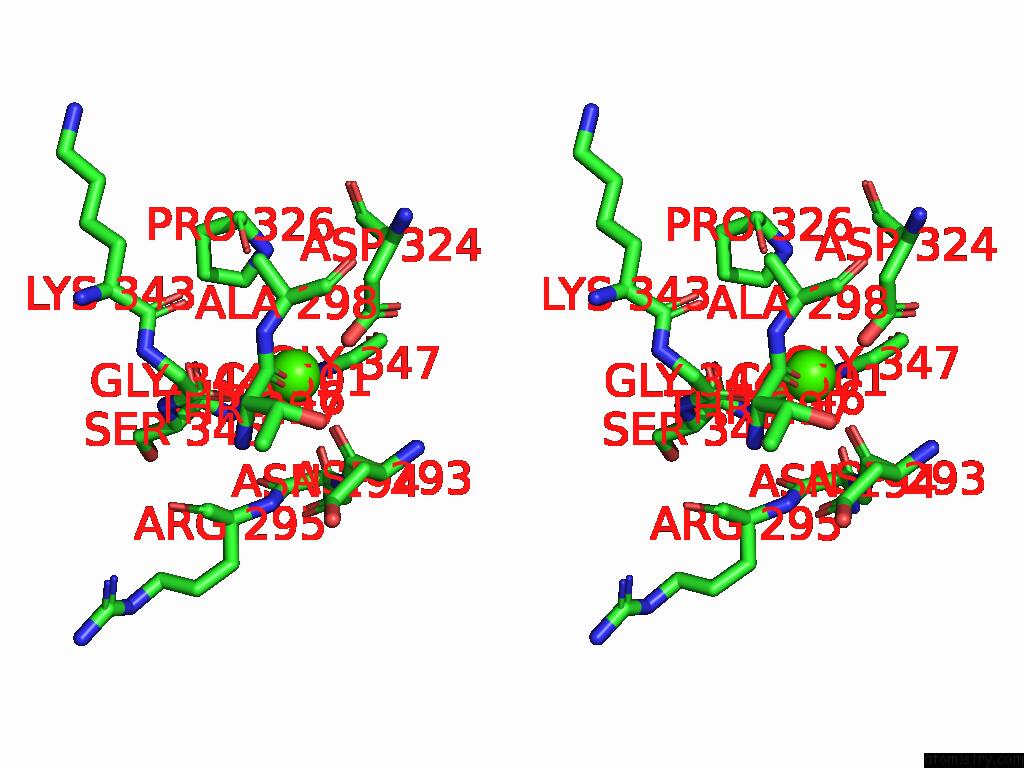
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 3 of Cryo-Em Structure of DA03E17 Fab in Complex with Influenza Virus Neuraminidase From B/Colorado/06/2017 within 5.0Å range:
|
Calcium binding site 4 out of 4 in 9cyh
Go back to
Calcium binding site 4 out
of 4 in the Cryo-Em Structure of DA03E17 Fab in Complex with Influenza Virus Neuraminidase From B/Colorado/06/2017
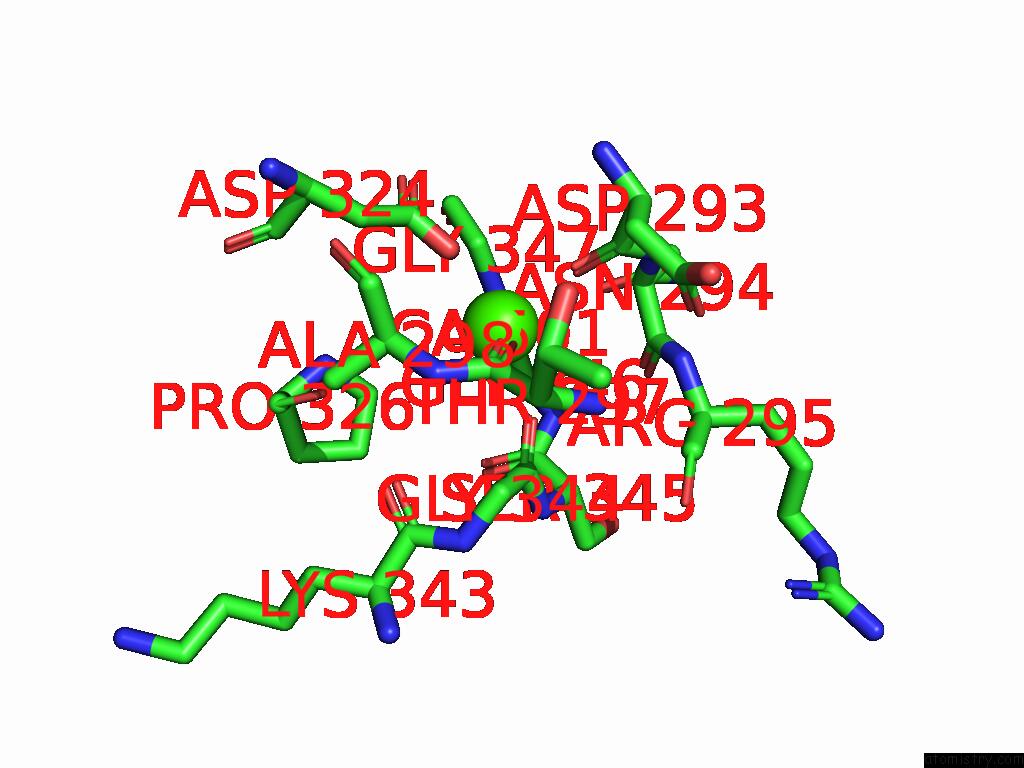
Mono view
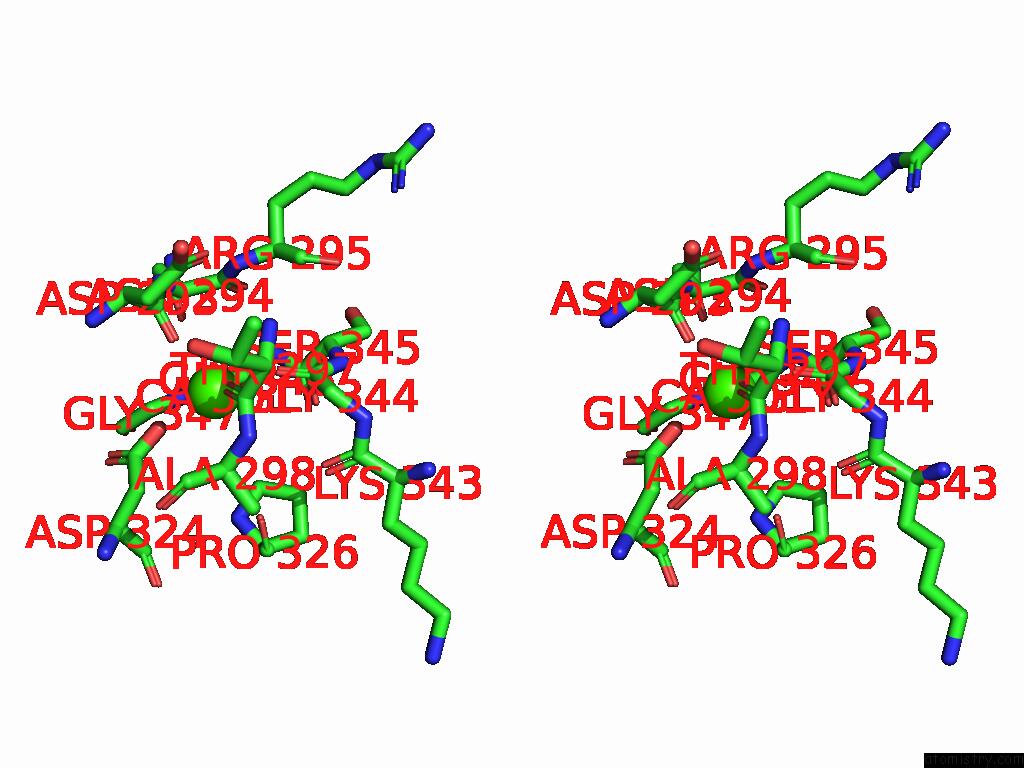
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 4 of Cryo-Em Structure of DA03E17 Fab in Complex with Influenza Virus Neuraminidase From B/Colorado/06/2017 within 5.0Å range:
|
Reference:
G.Jo,
S.Yamayoshi,
K.M.Ma,
O.Swanson,
J.L.Torres,
J.A.Ferguson,
M.L.Fernandez-Quintero,
J.Huang,
J.Copps,
A.J.Rodriguez,
J.M.Steichen,
Y.Kawaoka,
J.Han,
A.B.Ward.
Structural Basis of Broad Protection Against Influenza Virus By Human Antibodies Targeting the Neuraminidase Active Site Via A Recurring Motif in Cdr H3. Nat Commun V. 16 7067 2025.
ISSN: ESSN 2041-1723
PubMed: 40750588
DOI: 10.1038/S41467-025-62174-2
Page generated: Fri Aug 22 22:33:01 2025
ISSN: ESSN 2041-1723
PubMed: 40750588
DOI: 10.1038/S41467-025-62174-2
Last articles
K in 9NESK in 9PHG
K in 9NEI
K in 9NED
K in 9NEC
K in 9NEG
K in 9CWU
K in 9CVB
K in 9CVA
K in 9COM