Calcium »
PDB 9dho-9eit »
9eit »
Calcium in PDB 9eit: Ncs.1 Fab in Complex with N5 Na of A/Shorebird/Delaware Bay/309/2016 (DB16, H10N5) -- 4 Fabs
Enzymatic activity of Ncs.1 Fab in Complex with N5 Na of A/Shorebird/Delaware Bay/309/2016 (DB16, H10N5) -- 4 Fabs
All present enzymatic activity of Ncs.1 Fab in Complex with N5 Na of A/Shorebird/Delaware Bay/309/2016 (DB16, H10N5) -- 4 Fabs:
3.2.1.18;
3.2.1.18;
Calcium Binding Sites:
The binding sites of Calcium atom in the Ncs.1 Fab in Complex with N5 Na of A/Shorebird/Delaware Bay/309/2016 (DB16, H10N5) -- 4 Fabs
(pdb code 9eit). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 4 binding sites of Calcium where determined in the Ncs.1 Fab in Complex with N5 Na of A/Shorebird/Delaware Bay/309/2016 (DB16, H10N5) -- 4 Fabs, PDB code: 9eit:
Jump to Calcium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Calcium where determined in the Ncs.1 Fab in Complex with N5 Na of A/Shorebird/Delaware Bay/309/2016 (DB16, H10N5) -- 4 Fabs, PDB code: 9eit:
Jump to Calcium binding site number: 1; 2; 3; 4;
Calcium binding site 1 out of 4 in 9eit
Go back to
Calcium binding site 1 out
of 4 in the Ncs.1 Fab in Complex with N5 Na of A/Shorebird/Delaware Bay/309/2016 (DB16, H10N5) -- 4 Fabs
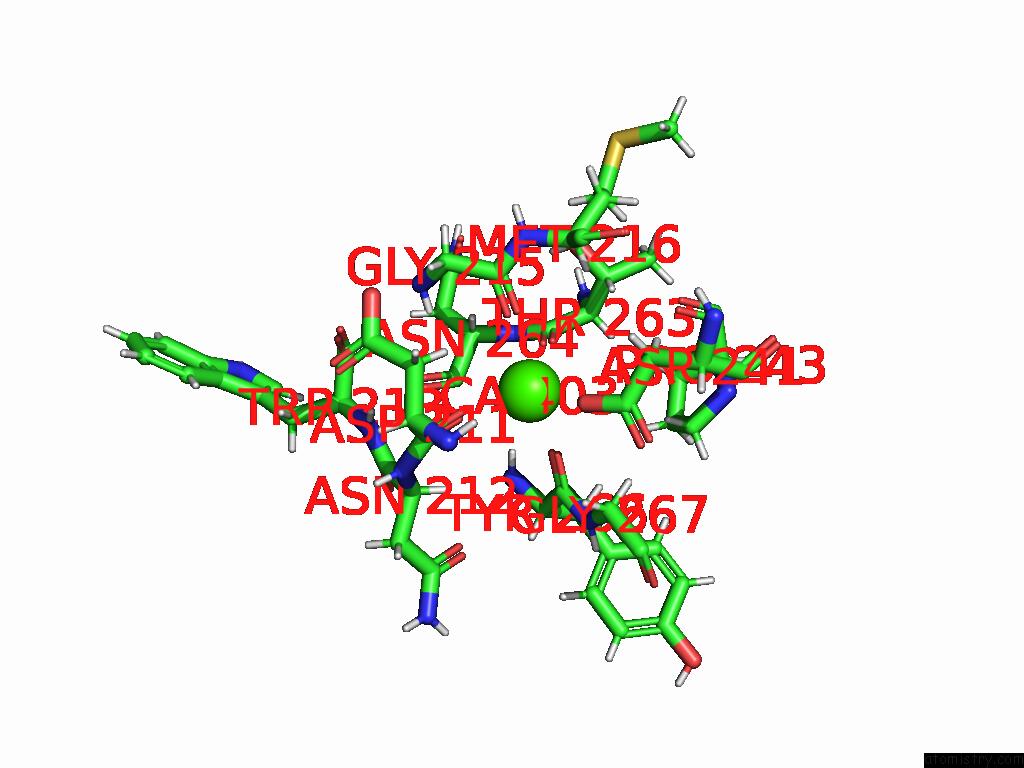
Mono view
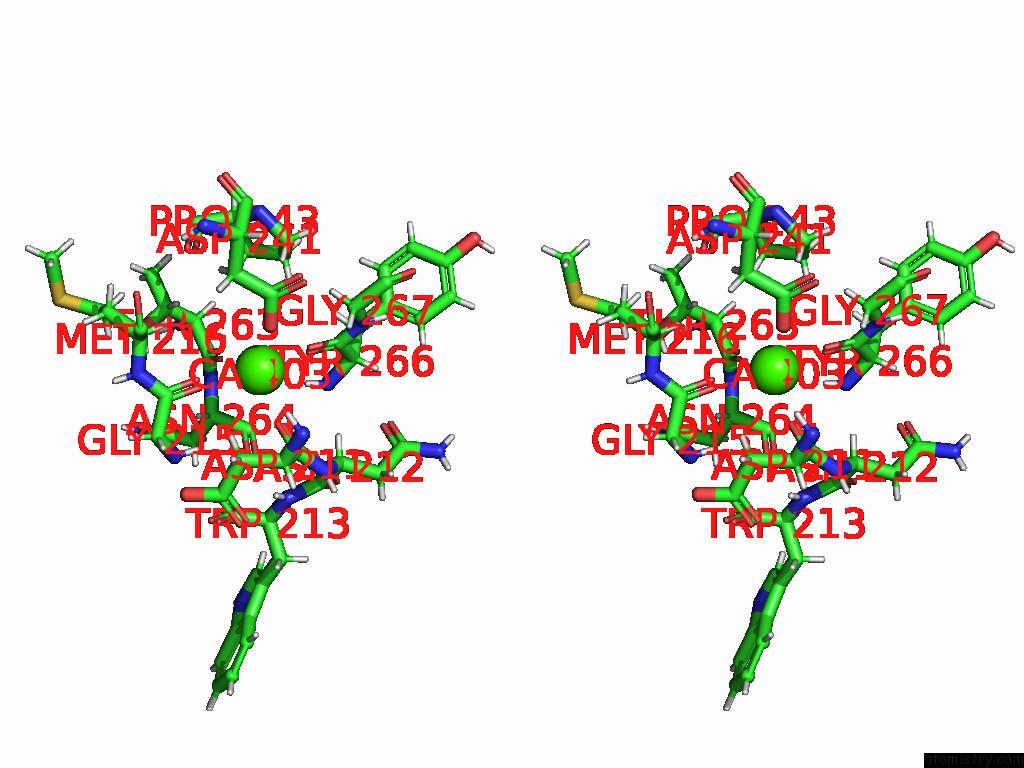
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Ncs.1 Fab in Complex with N5 Na of A/Shorebird/Delaware Bay/309/2016 (DB16, H10N5) -- 4 Fabs within 5.0Å range:
|
Calcium binding site 2 out of 4 in 9eit
Go back to
Calcium binding site 2 out
of 4 in the Ncs.1 Fab in Complex with N5 Na of A/Shorebird/Delaware Bay/309/2016 (DB16, H10N5) -- 4 Fabs
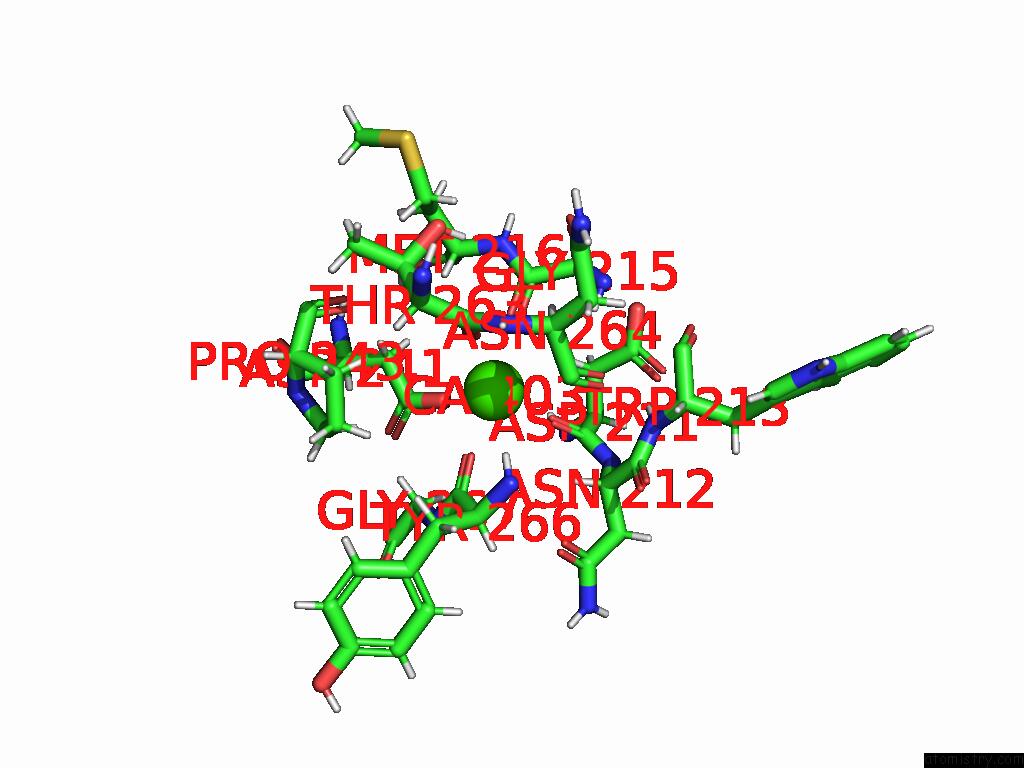
Mono view
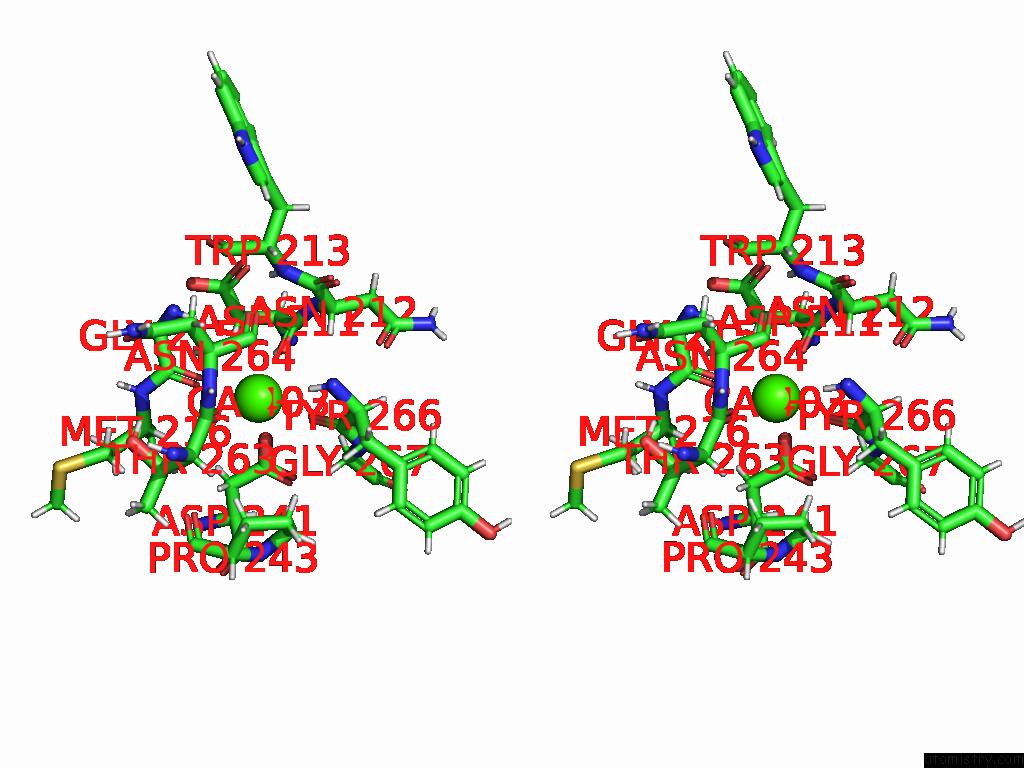
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Ncs.1 Fab in Complex with N5 Na of A/Shorebird/Delaware Bay/309/2016 (DB16, H10N5) -- 4 Fabs within 5.0Å range:
|
Calcium binding site 3 out of 4 in 9eit
Go back to
Calcium binding site 3 out
of 4 in the Ncs.1 Fab in Complex with N5 Na of A/Shorebird/Delaware Bay/309/2016 (DB16, H10N5) -- 4 Fabs
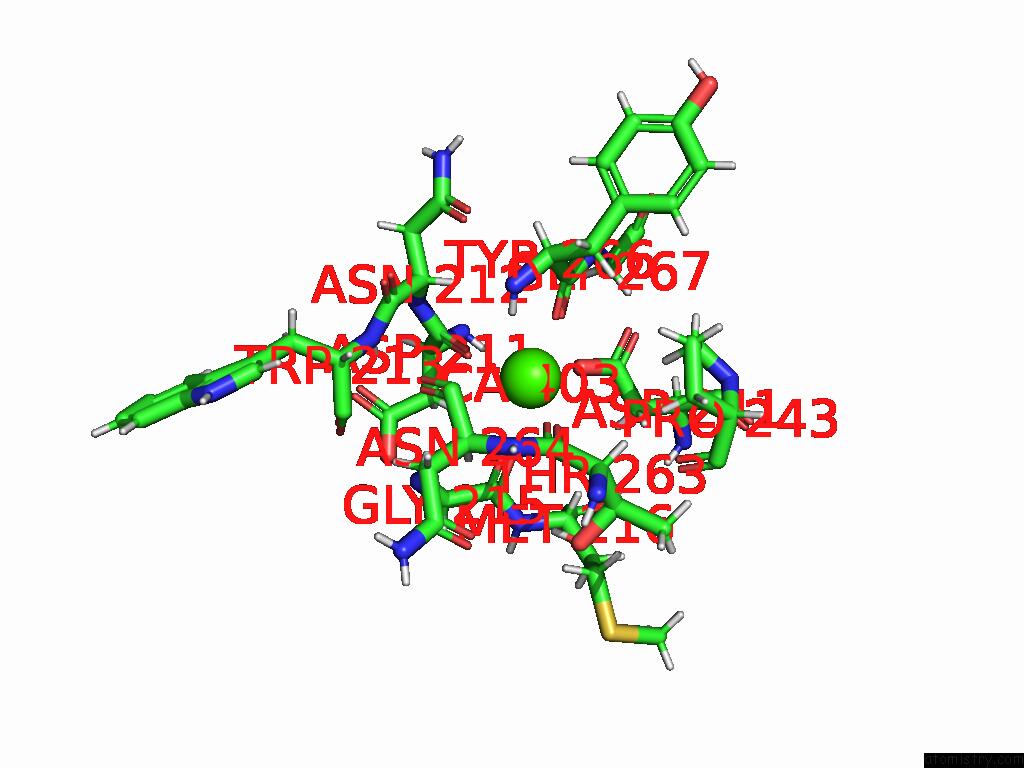
Mono view
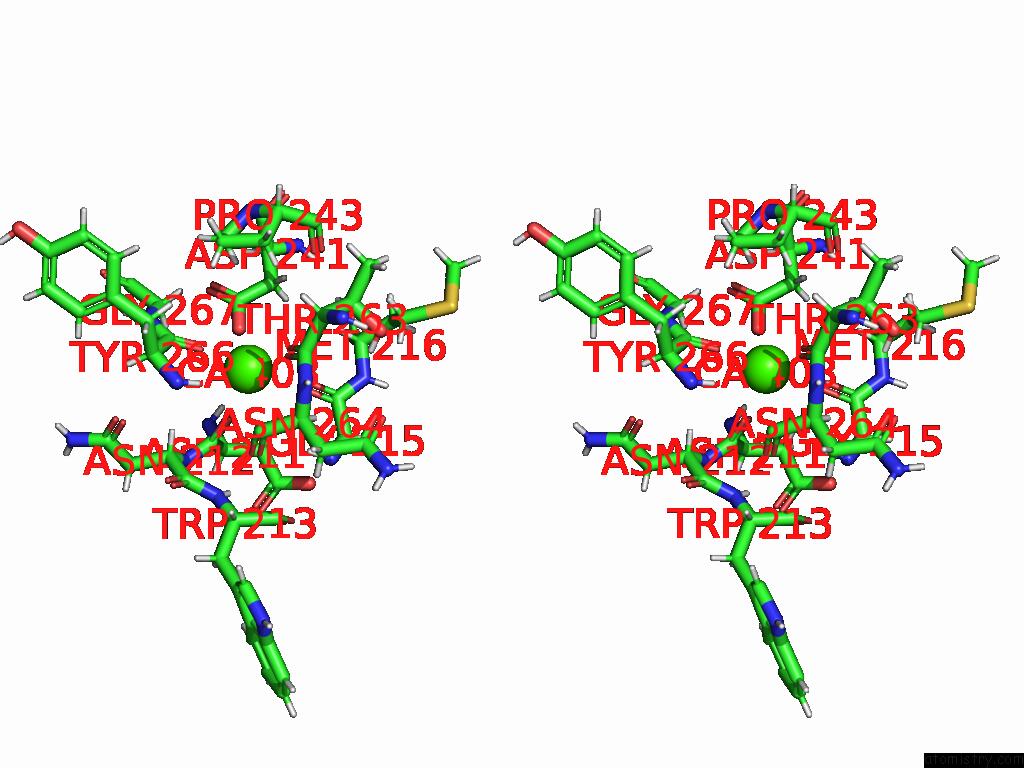
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 3 of Ncs.1 Fab in Complex with N5 Na of A/Shorebird/Delaware Bay/309/2016 (DB16, H10N5) -- 4 Fabs within 5.0Å range:
|
Calcium binding site 4 out of 4 in 9eit
Go back to
Calcium binding site 4 out
of 4 in the Ncs.1 Fab in Complex with N5 Na of A/Shorebird/Delaware Bay/309/2016 (DB16, H10N5) -- 4 Fabs
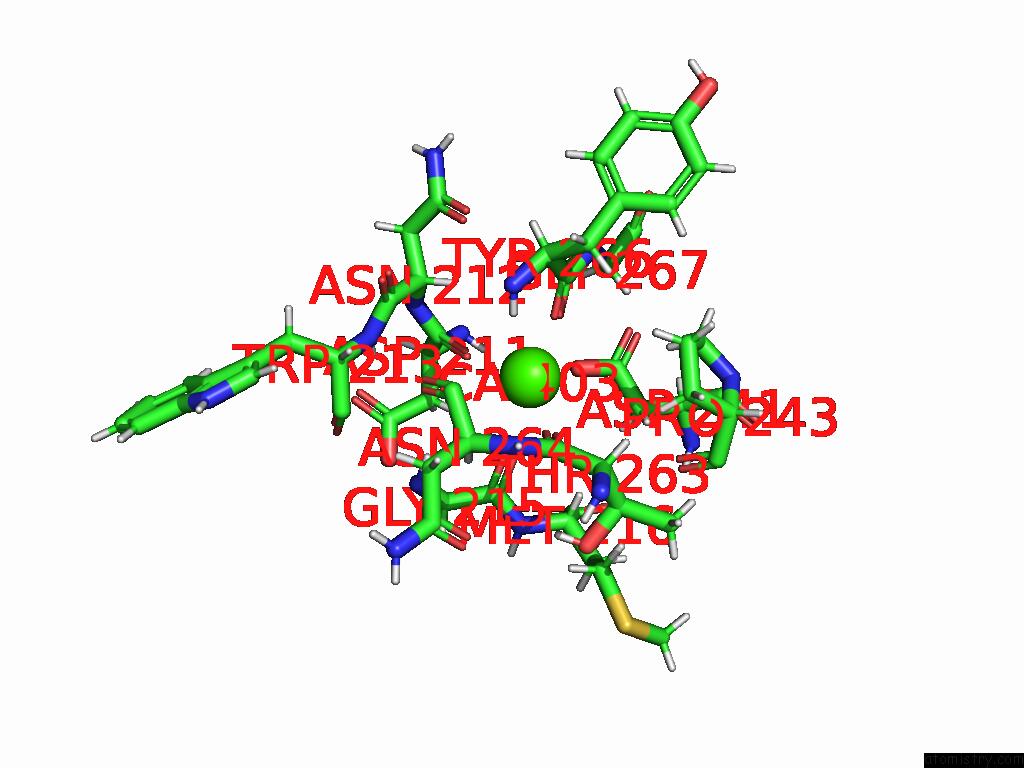
Mono view
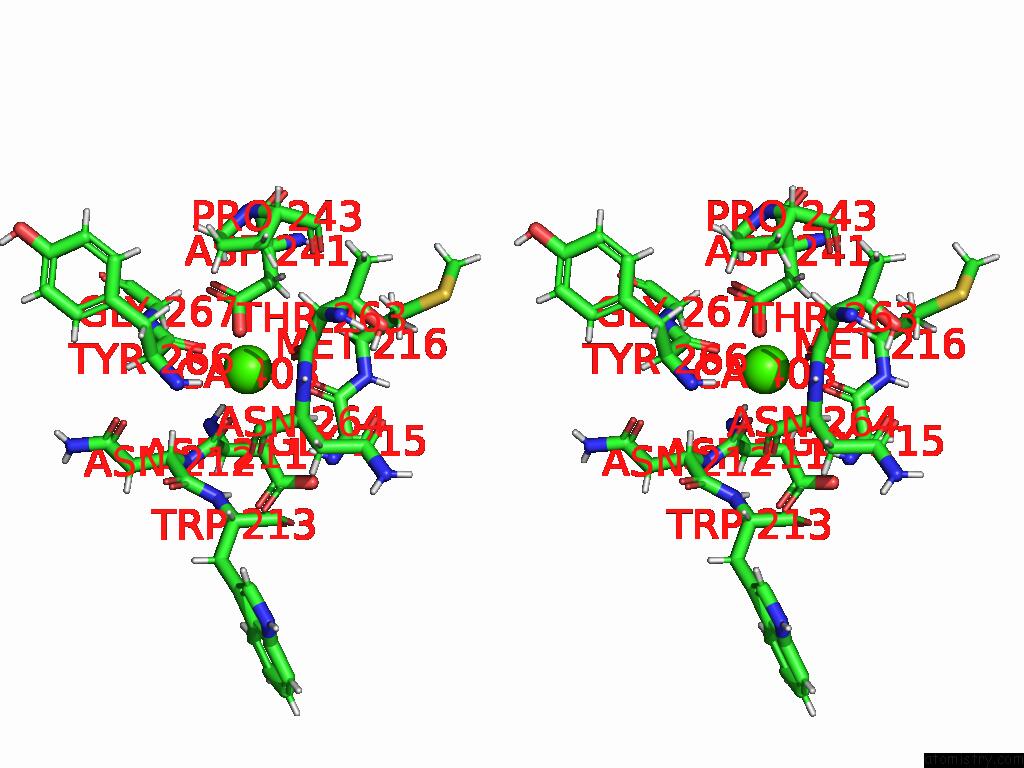
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 4 of Ncs.1 Fab in Complex with N5 Na of A/Shorebird/Delaware Bay/309/2016 (DB16, H10N5) -- 4 Fabs within 5.0Å range:
|
Reference:
J.Lederhofer,
A.J.Borst,
L.Nguyen,
R.A.Gillespie,
C.J.Williams,
E.L.Walker,
J.E.Raab,
C.Yap,
D.Ellis,
A.Creanga,
H.X.Tan,
T.H.T.Do,
M.Ravichandran,
A.B.Mcdermott,
V.Le Sage,
S.F.Andrews,
B.S.Graham,
A.K.Wheatley,
D.S.Reed,
N.P.King,
M.Kanekiyo.
Structural Convergence and Water-Mediated Substrate Mimicry Enable Broad Neuraminidase Inhibition By Human Antibodies. Nat Commun V. 16 7068 2025.
ISSN: ESSN 2041-1723
PubMed: 40750617
DOI: 10.1038/S41467-025-62339-Z
Page generated: Fri Aug 22 22:34:47 2025
ISSN: ESSN 2041-1723
PubMed: 40750617
DOI: 10.1038/S41467-025-62339-Z
Last articles
Mn in 9LJUMn in 9LJW
Mn in 9LJS
Mn in 9LJR
Mn in 9LJT
Mn in 9LJV
Mg in 9UA2
Mg in 9R96
Mg in 9VM1
Mg in 9P01