Calcium »
PDB 9eje-9f8x »
9eje »
Calcium in PDB 9eje: Ncs.1 Fab in Complex with N5 Na of A/Shorebird/Delaware Bay/309/2016 (DB16, H10N5) -- 3 Fabs
Enzymatic activity of Ncs.1 Fab in Complex with N5 Na of A/Shorebird/Delaware Bay/309/2016 (DB16, H10N5) -- 3 Fabs
All present enzymatic activity of Ncs.1 Fab in Complex with N5 Na of A/Shorebird/Delaware Bay/309/2016 (DB16, H10N5) -- 3 Fabs:
3.2.1.18;
3.2.1.18;
Calcium Binding Sites:
The binding sites of Calcium atom in the Ncs.1 Fab in Complex with N5 Na of A/Shorebird/Delaware Bay/309/2016 (DB16, H10N5) -- 3 Fabs
(pdb code 9eje). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 4 binding sites of Calcium where determined in the Ncs.1 Fab in Complex with N5 Na of A/Shorebird/Delaware Bay/309/2016 (DB16, H10N5) -- 3 Fabs, PDB code: 9eje:
Jump to Calcium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Calcium where determined in the Ncs.1 Fab in Complex with N5 Na of A/Shorebird/Delaware Bay/309/2016 (DB16, H10N5) -- 3 Fabs, PDB code: 9eje:
Jump to Calcium binding site number: 1; 2; 3; 4;
Calcium binding site 1 out of 4 in 9eje
Go back to
Calcium binding site 1 out
of 4 in the Ncs.1 Fab in Complex with N5 Na of A/Shorebird/Delaware Bay/309/2016 (DB16, H10N5) -- 3 Fabs
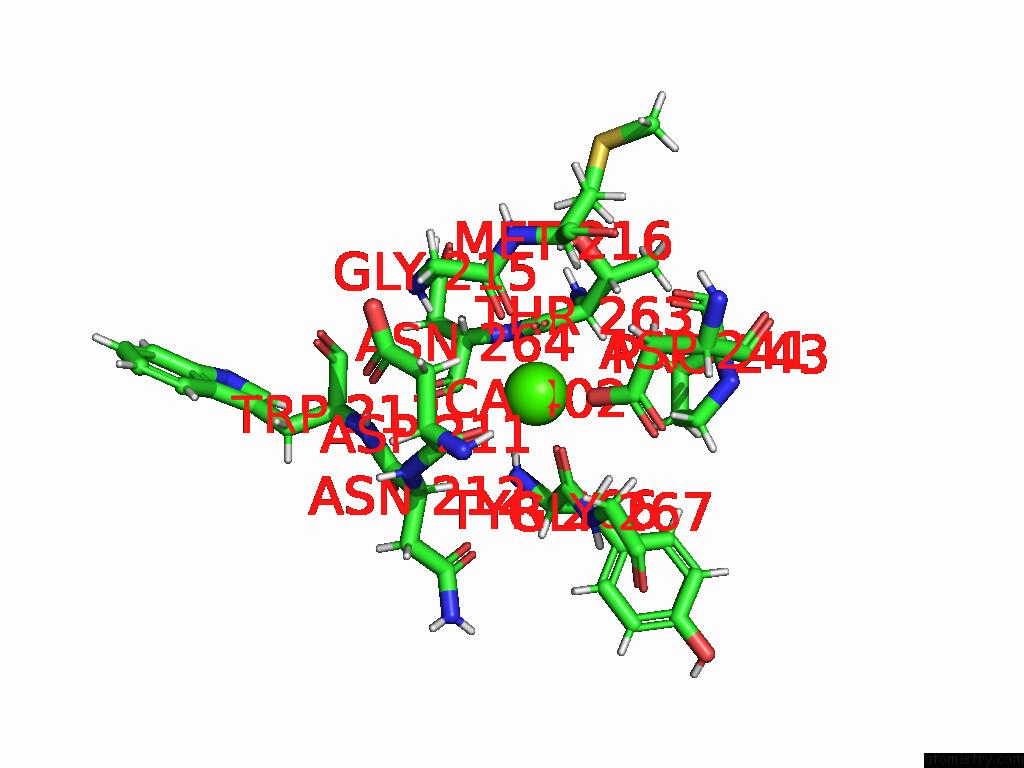
Mono view
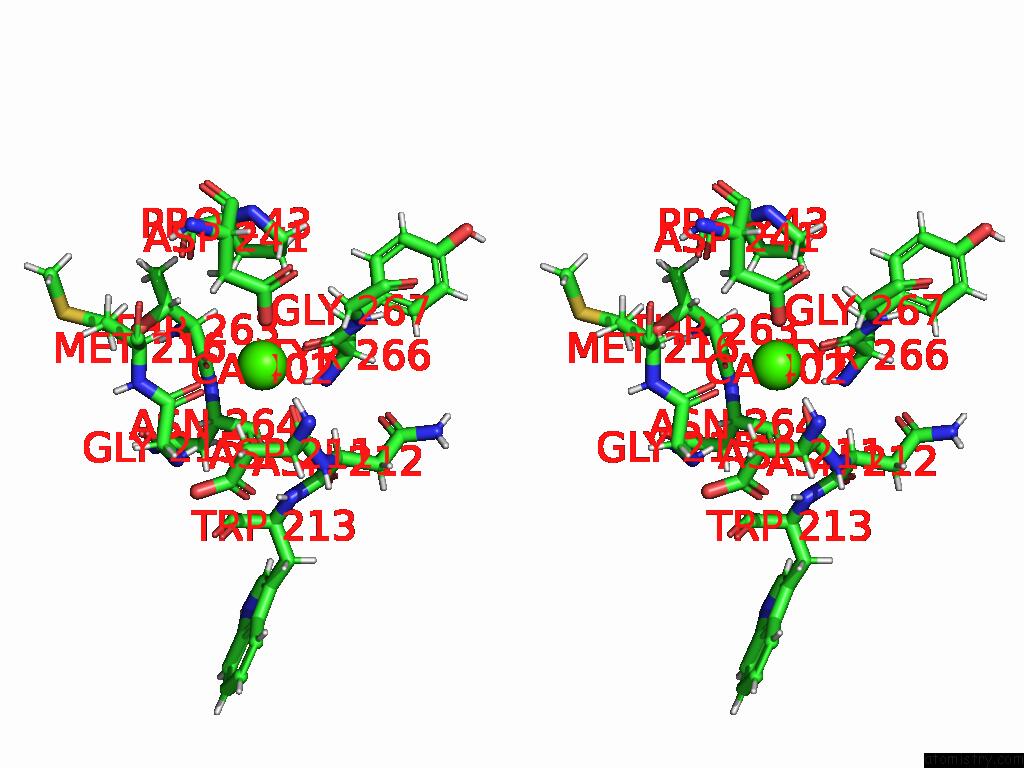
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Ncs.1 Fab in Complex with N5 Na of A/Shorebird/Delaware Bay/309/2016 (DB16, H10N5) -- 3 Fabs within 5.0Å range:
|
Calcium binding site 2 out of 4 in 9eje
Go back to
Calcium binding site 2 out
of 4 in the Ncs.1 Fab in Complex with N5 Na of A/Shorebird/Delaware Bay/309/2016 (DB16, H10N5) -- 3 Fabs
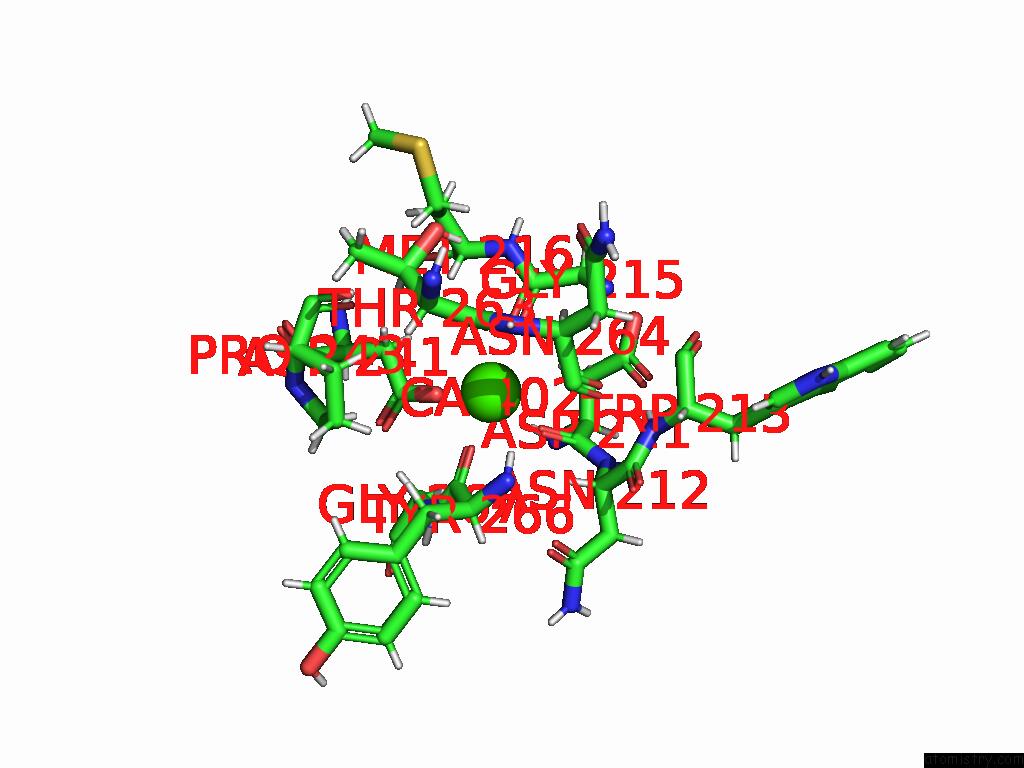
Mono view
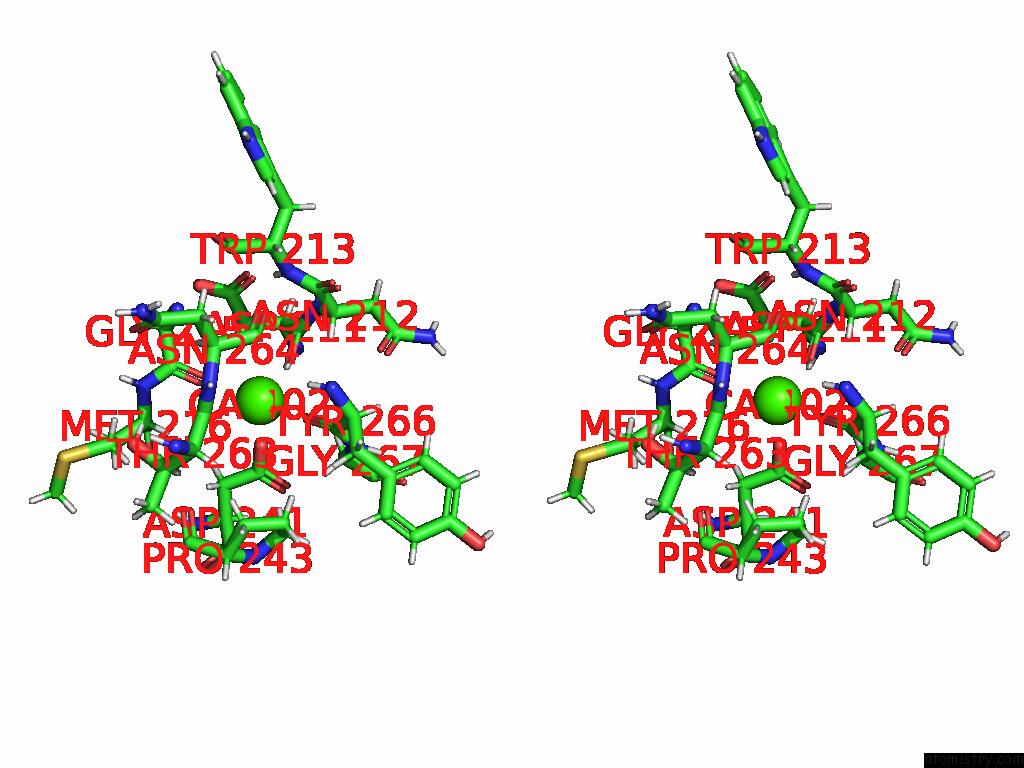
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Ncs.1 Fab in Complex with N5 Na of A/Shorebird/Delaware Bay/309/2016 (DB16, H10N5) -- 3 Fabs within 5.0Å range:
|
Calcium binding site 3 out of 4 in 9eje
Go back to
Calcium binding site 3 out
of 4 in the Ncs.1 Fab in Complex with N5 Na of A/Shorebird/Delaware Bay/309/2016 (DB16, H10N5) -- 3 Fabs
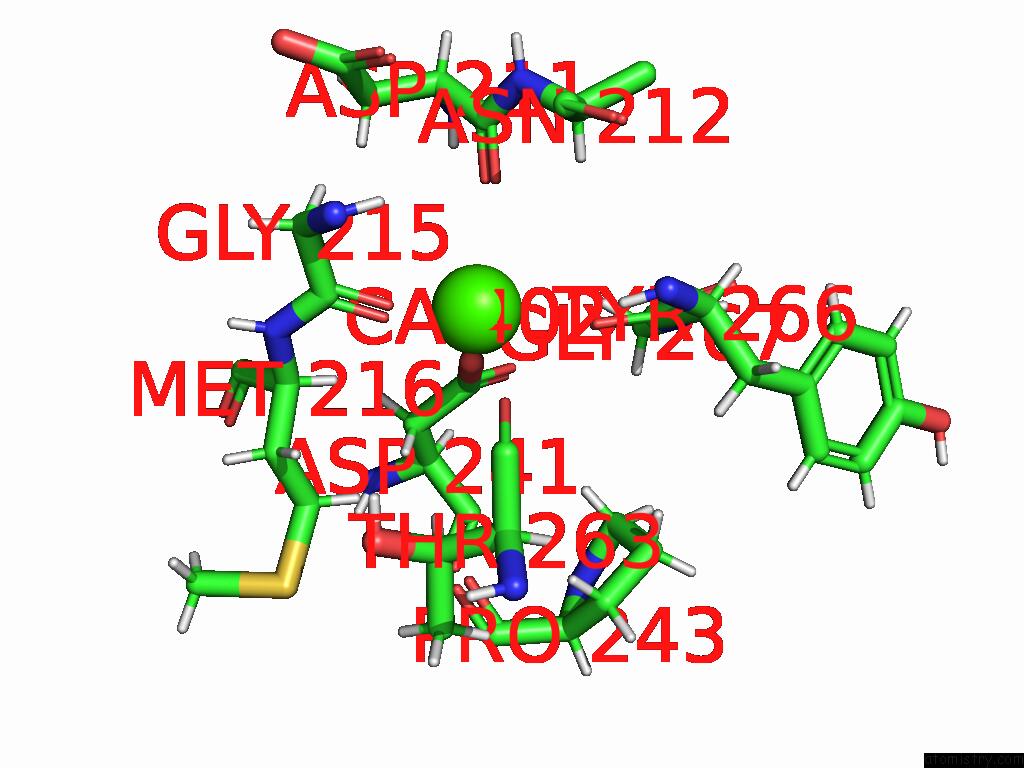
Mono view
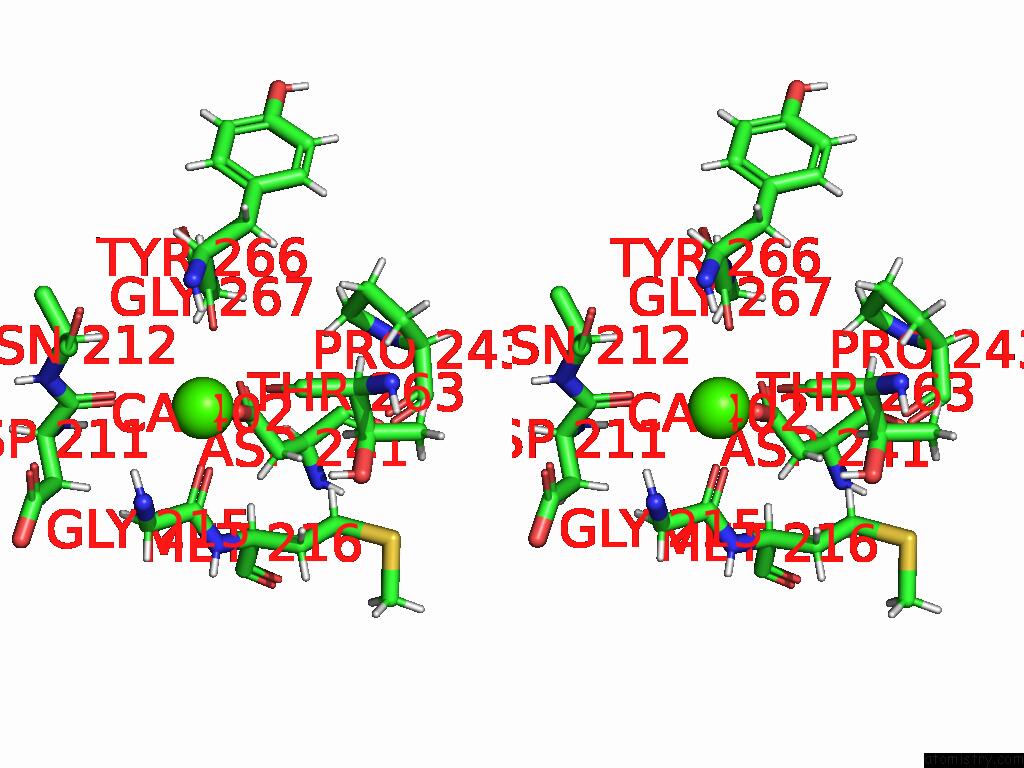
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 3 of Ncs.1 Fab in Complex with N5 Na of A/Shorebird/Delaware Bay/309/2016 (DB16, H10N5) -- 3 Fabs within 5.0Å range:
|
Calcium binding site 4 out of 4 in 9eje
Go back to
Calcium binding site 4 out
of 4 in the Ncs.1 Fab in Complex with N5 Na of A/Shorebird/Delaware Bay/309/2016 (DB16, H10N5) -- 3 Fabs
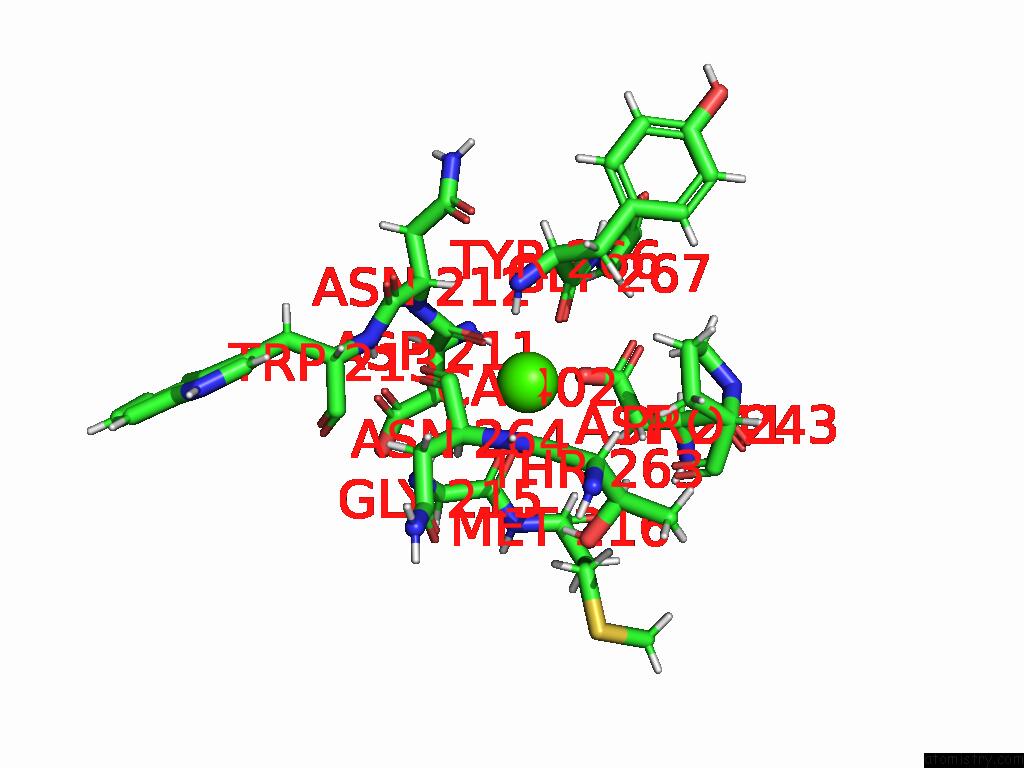
Mono view
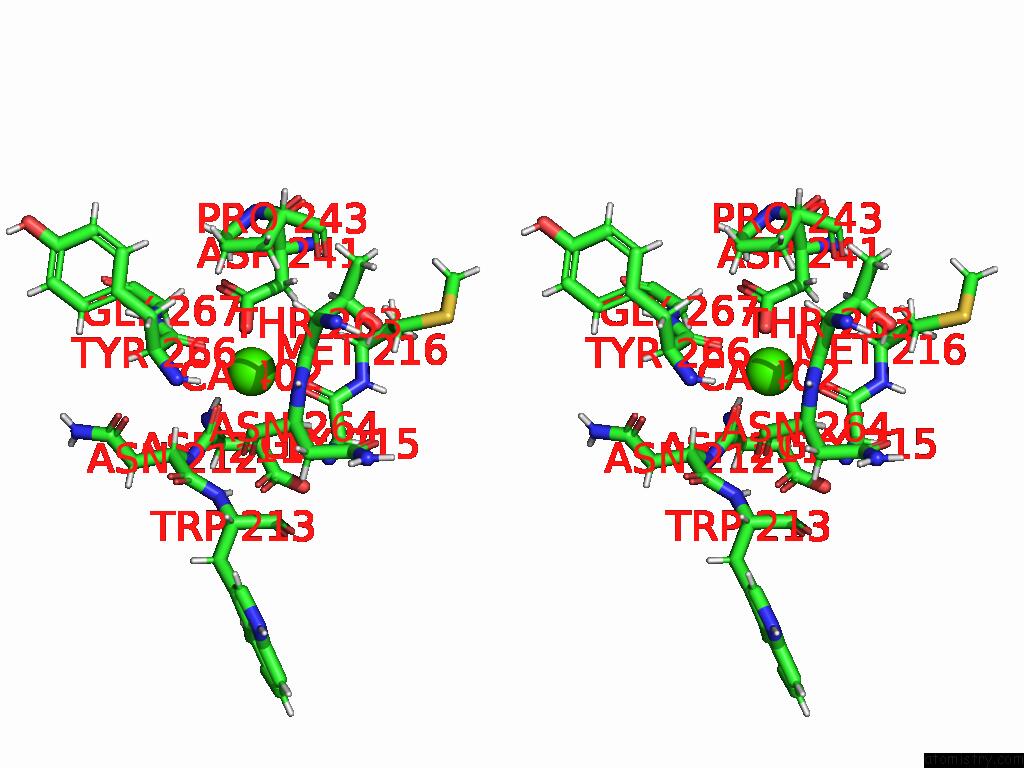
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 4 of Ncs.1 Fab in Complex with N5 Na of A/Shorebird/Delaware Bay/309/2016 (DB16, H10N5) -- 3 Fabs within 5.0Å range:
|
Reference:
J.Lederhofer,
A.J.Borst,
L.Nguyen,
R.A.Gillespie,
C.J.Williams,
E.L.Walker,
J.E.Raab,
C.Yap,
D.Ellis,
A.Creanga,
H.X.Tan,
T.H.T.Do,
M.Ravichandran,
A.B.Mcdermott,
V.Le Sage,
S.F.Andrews,
B.S.Graham,
A.K.Wheatley,
D.S.Reed,
N.P.King,
M.Kanekiyo.
Structural Convergence and Water-Mediated Substrate Mimicry Enable Broad Neuraminidase Inhibition By Human Antibodies. Nat Commun V. 16 7068 2025.
ISSN: ESSN 2041-1723
PubMed: 40750617
DOI: 10.1038/S41467-025-62339-Z
Page generated: Fri Aug 22 22:35:03 2025
ISSN: ESSN 2041-1723
PubMed: 40750617
DOI: 10.1038/S41467-025-62339-Z
Last articles
K in 9NESK in 9PHG
K in 9NEI
K in 9NED
K in 9NEC
K in 9NEG
K in 9CWU
K in 9CVB
K in 9CVA
K in 9COM