Calcium »
PDB 1cxk-1dds »
1daq »
Calcium in PDB 1daq: Solution Structure of the Type I Dockerin Domain From the Clostridium Thermocellum Cellulosome (Minimized Average Structure)
Enzymatic activity of Solution Structure of the Type I Dockerin Domain From the Clostridium Thermocellum Cellulosome (Minimized Average Structure)
All present enzymatic activity of Solution Structure of the Type I Dockerin Domain From the Clostridium Thermocellum Cellulosome (Minimized Average Structure):
3.2.1.4;
3.2.1.4;
Calcium Binding Sites:
The binding sites of Calcium atom in the Solution Structure of the Type I Dockerin Domain From the Clostridium Thermocellum Cellulosome (Minimized Average Structure)
(pdb code 1daq). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 2 binding sites of Calcium where determined in the Solution Structure of the Type I Dockerin Domain From the Clostridium Thermocellum Cellulosome (Minimized Average Structure), PDB code: 1daq:
Jump to Calcium binding site number: 1; 2;
In total 2 binding sites of Calcium where determined in the Solution Structure of the Type I Dockerin Domain From the Clostridium Thermocellum Cellulosome (Minimized Average Structure), PDB code: 1daq:
Jump to Calcium binding site number: 1; 2;
Calcium binding site 1 out of 2 in 1daq
Go back to
Calcium binding site 1 out
of 2 in the Solution Structure of the Type I Dockerin Domain From the Clostridium Thermocellum Cellulosome (Minimized Average Structure)

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Solution Structure of the Type I Dockerin Domain From the Clostridium Thermocellum Cellulosome (Minimized Average Structure) within 5.0Å range:
|
Calcium binding site 2 out of 2 in 1daq
Go back to
Calcium binding site 2 out
of 2 in the Solution Structure of the Type I Dockerin Domain From the Clostridium Thermocellum Cellulosome (Minimized Average Structure)
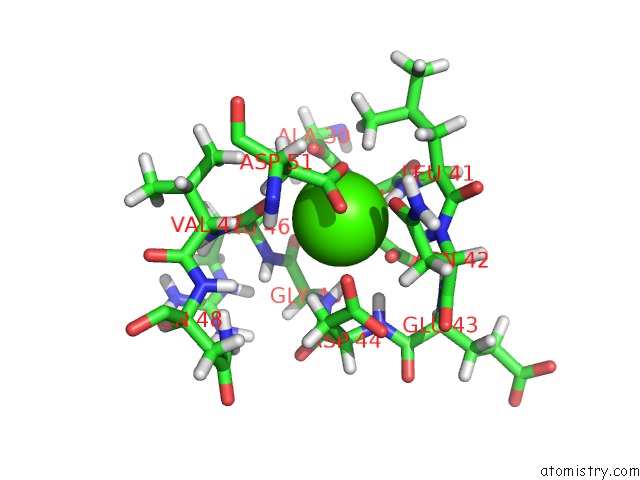
Mono view
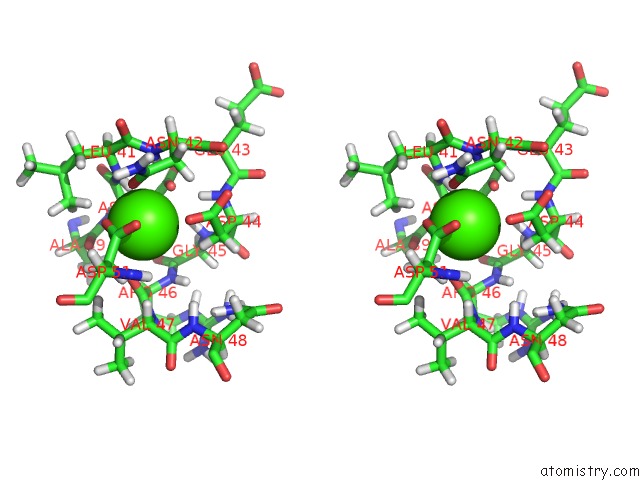
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Solution Structure of the Type I Dockerin Domain From the Clostridium Thermocellum Cellulosome (Minimized Average Structure) within 5.0Å range:
|
Reference:
B.L.Lytle,
B.F.Volkman,
W.M.Westler,
M.P.Heckman,
J.H.Wu.
Solution Structure of A Type I Dockerin Domain, A Novel Prokaryotic, Extracellular Calcium-Binding Domain. J.Mol.Biol. V. 307 745 2001.
ISSN: ISSN 0022-2836
PubMed: 11273698
DOI: 10.1006/JMBI.2001.4522
Page generated: Mon Jul 7 14:22:05 2025
ISSN: ISSN 0022-2836
PubMed: 11273698
DOI: 10.1006/JMBI.2001.4522
Last articles
Mg in 4DV2Mg in 4DV0
Mg in 4DV1
Mg in 4DUZ
Mg in 4DUY
Mg in 4DR7
Mg in 4DR6
Mg in 4DR5
Mg in 4DUX
Mg in 4DUW