Calcium »
PDB 1lu1-1m8v »
1m5x »
Calcium in PDB 1m5x: Crystal Structure of the Homing Endonuclease I-Msoi Bound to Its Dna Substrate
Protein crystallography data
The structure of Crystal Structure of the Homing Endonuclease I-Msoi Bound to Its Dna Substrate, PDB code: 1m5x
was solved by
B.Chevalier,
M.Turmel,
C.Lemieux,
R.J.Monnat,
B.L.Stoddard,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 19.96 / 2.25 |
| Space group | P 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 41.510, 42.210, 71.320, 73.34, 73.17, 71.09 |
| R / Rfree (%) | 21.9 / 25.5 |
Calcium Binding Sites:
The binding sites of Calcium atom in the Crystal Structure of the Homing Endonuclease I-Msoi Bound to Its Dna Substrate
(pdb code 1m5x). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 3 binding sites of Calcium where determined in the Crystal Structure of the Homing Endonuclease I-Msoi Bound to Its Dna Substrate, PDB code: 1m5x:
Jump to Calcium binding site number: 1; 2; 3;
In total 3 binding sites of Calcium where determined in the Crystal Structure of the Homing Endonuclease I-Msoi Bound to Its Dna Substrate, PDB code: 1m5x:
Jump to Calcium binding site number: 1; 2; 3;
Calcium binding site 1 out of 3 in 1m5x
Go back to
Calcium binding site 1 out
of 3 in the Crystal Structure of the Homing Endonuclease I-Msoi Bound to Its Dna Substrate
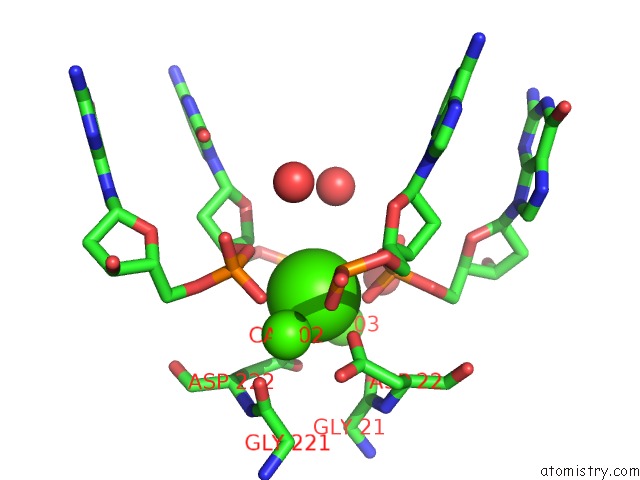
Mono view
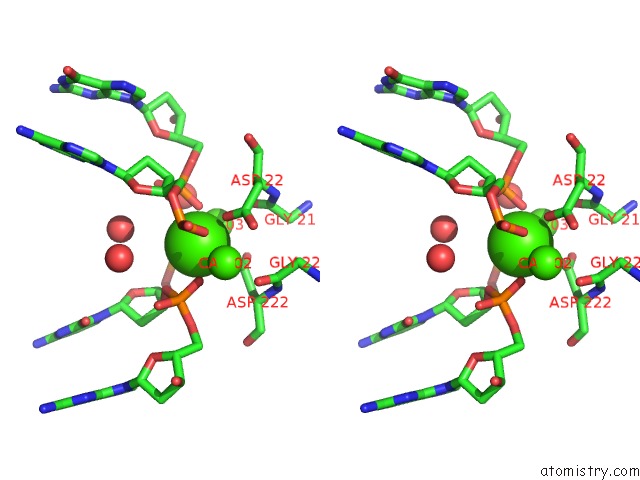
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Crystal Structure of the Homing Endonuclease I-Msoi Bound to Its Dna Substrate within 5.0Å range:
|
Calcium binding site 2 out of 3 in 1m5x
Go back to
Calcium binding site 2 out
of 3 in the Crystal Structure of the Homing Endonuclease I-Msoi Bound to Its Dna Substrate
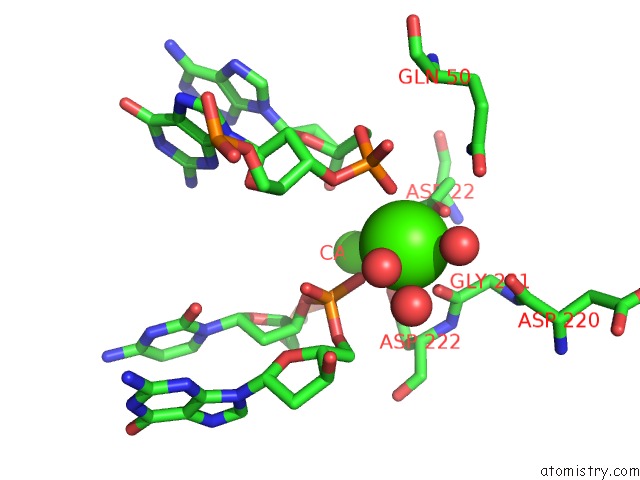
Mono view
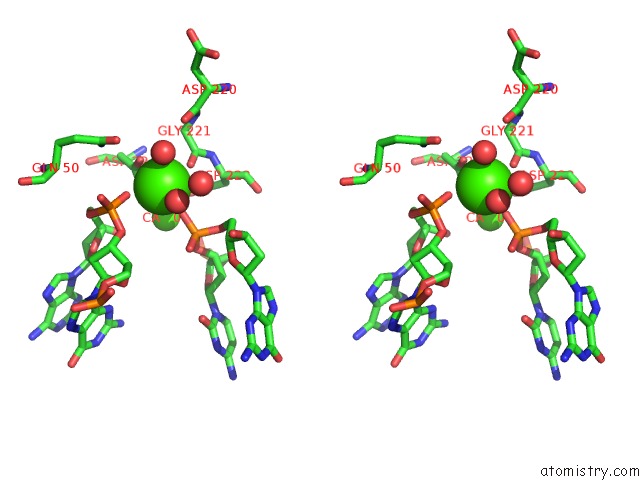
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Crystal Structure of the Homing Endonuclease I-Msoi Bound to Its Dna Substrate within 5.0Å range:
|
Calcium binding site 3 out of 3 in 1m5x
Go back to
Calcium binding site 3 out
of 3 in the Crystal Structure of the Homing Endonuclease I-Msoi Bound to Its Dna Substrate

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 3 of Crystal Structure of the Homing Endonuclease I-Msoi Bound to Its Dna Substrate within 5.0Å range:
|
Reference:
B.Chevalier,
M.Turmel,
C.Lemieux,
R.J.Monnat,
B.L.Stoddard.
Flexible Dna Target Site Recognition By Divergent Homing Endonuclease Isoschizomers I-Crei and I-Msoi J.Mol.Biol. V. 329 253 2003.
ISSN: ISSN 0022-2836
PubMed: 12758074
DOI: 10.1016/S0022-2836(03)00447-9
Page generated: Mon Jul 7 17:13:01 2025
ISSN: ISSN 0022-2836
PubMed: 12758074
DOI: 10.1016/S0022-2836(03)00447-9
Last articles
Ir in 5OD5Ir in 5JML
Ir in 5T83
Ir in 5K7D
Ir in 5E1U
Ir in 5JMG
Ir in 5HQO
Ir in 5E2D
Ir in 4R0D
Ir in 4Y1N