Calcium »
PDB 1mts-1n7d »
1mux »
Calcium in PDB 1mux: Solution uc(Nmr) Structure of Calmodulin/W-7 Complex: the Basis of Diversity in Molecular Recognition, 30 Structures
Other elements in 1mux:
The structure of Solution uc(Nmr) Structure of Calmodulin/W-7 Complex: the Basis of Diversity in Molecular Recognition, 30 Structures also contains other interesting chemical elements:
| Chlorine | (Cl) | 60 atoms |
Calcium Binding Sites:
The binding sites of Calcium atom in the Solution uc(Nmr) Structure of Calmodulin/W-7 Complex: the Basis of Diversity in Molecular Recognition, 30 Structures
(pdb code 1mux). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 4 binding sites of Calcium where determined in the Solution uc(Nmr) Structure of Calmodulin/W-7 Complex: the Basis of Diversity in Molecular Recognition, 30 Structures, PDB code: 1mux:
Jump to Calcium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Calcium where determined in the Solution uc(Nmr) Structure of Calmodulin/W-7 Complex: the Basis of Diversity in Molecular Recognition, 30 Structures, PDB code: 1mux:
Jump to Calcium binding site number: 1; 2; 3; 4;
Calcium binding site 1 out of 4 in 1mux
Go back to
Calcium binding site 1 out
of 4 in the Solution uc(Nmr) Structure of Calmodulin/W-7 Complex: the Basis of Diversity in Molecular Recognition, 30 Structures
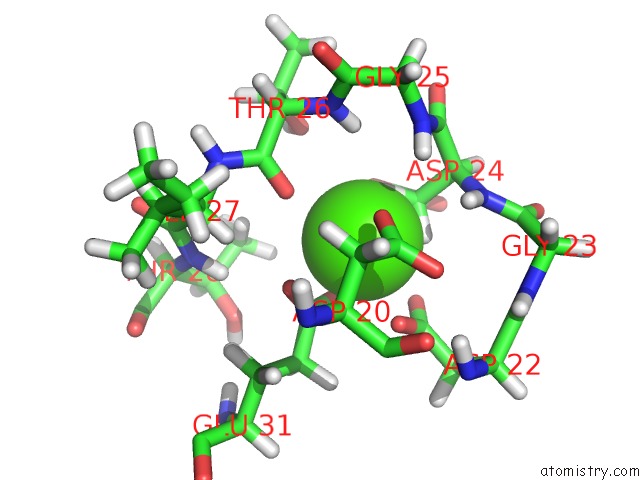
Mono view
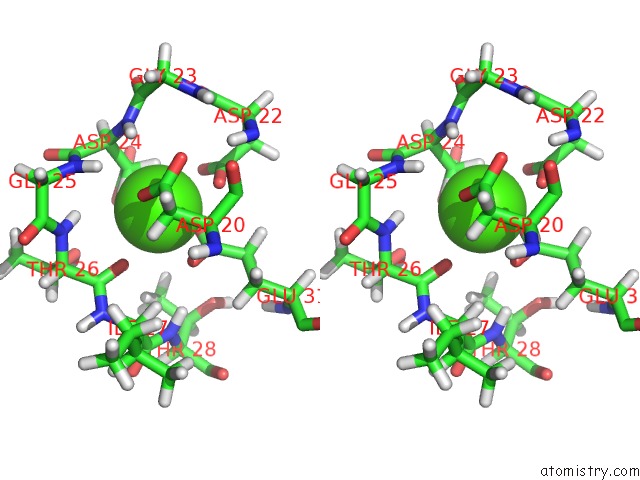
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Solution uc(Nmr) Structure of Calmodulin/W-7 Complex: the Basis of Diversity in Molecular Recognition, 30 Structures within 5.0Å range:
|
Calcium binding site 2 out of 4 in 1mux
Go back to
Calcium binding site 2 out
of 4 in the Solution uc(Nmr) Structure of Calmodulin/W-7 Complex: the Basis of Diversity in Molecular Recognition, 30 Structures
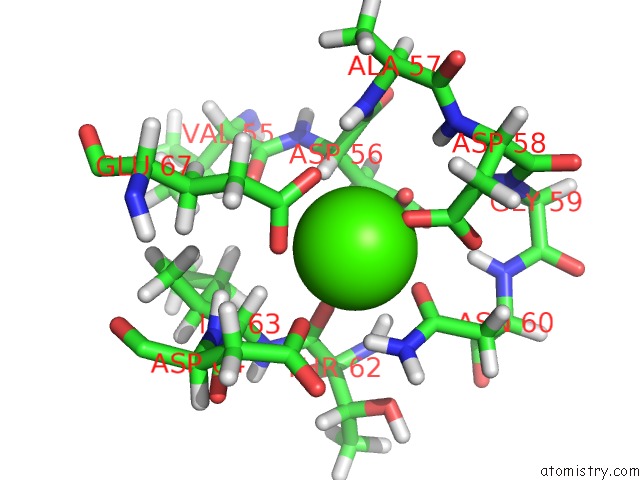
Mono view
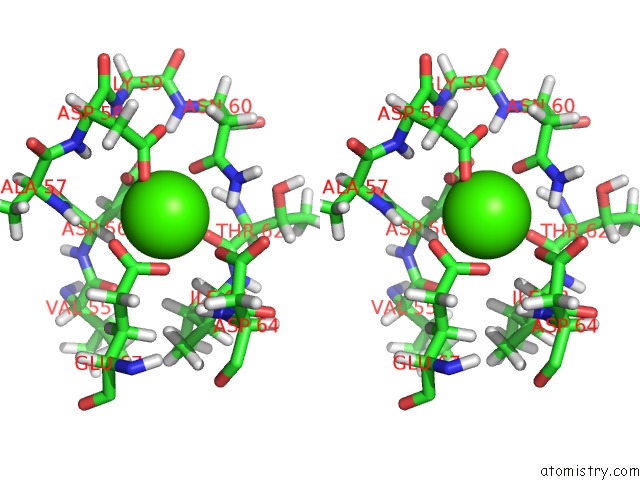
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Solution uc(Nmr) Structure of Calmodulin/W-7 Complex: the Basis of Diversity in Molecular Recognition, 30 Structures within 5.0Å range:
|
Calcium binding site 3 out of 4 in 1mux
Go back to
Calcium binding site 3 out
of 4 in the Solution uc(Nmr) Structure of Calmodulin/W-7 Complex: the Basis of Diversity in Molecular Recognition, 30 Structures
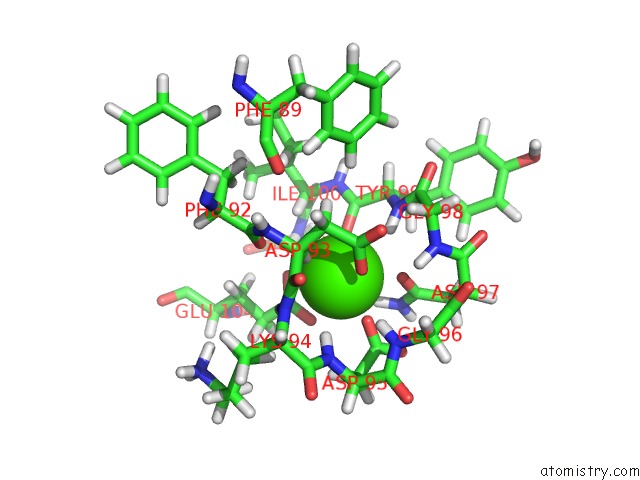
Mono view
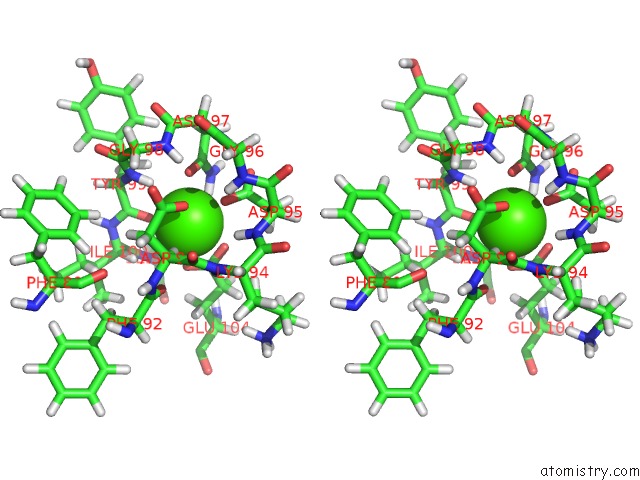
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 3 of Solution uc(Nmr) Structure of Calmodulin/W-7 Complex: the Basis of Diversity in Molecular Recognition, 30 Structures within 5.0Å range:
|
Calcium binding site 4 out of 4 in 1mux
Go back to
Calcium binding site 4 out
of 4 in the Solution uc(Nmr) Structure of Calmodulin/W-7 Complex: the Basis of Diversity in Molecular Recognition, 30 Structures
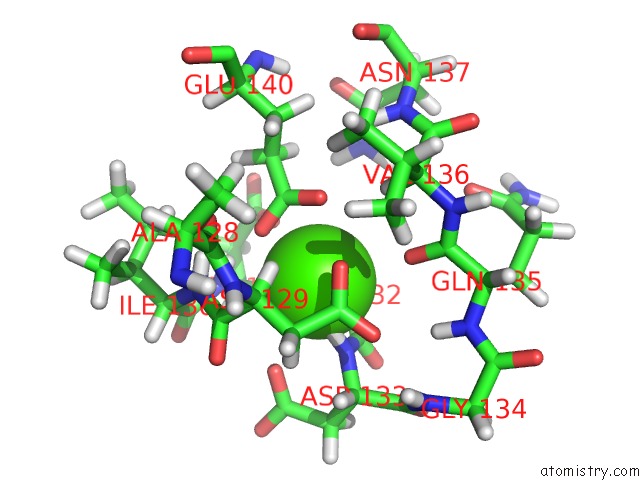
Mono view
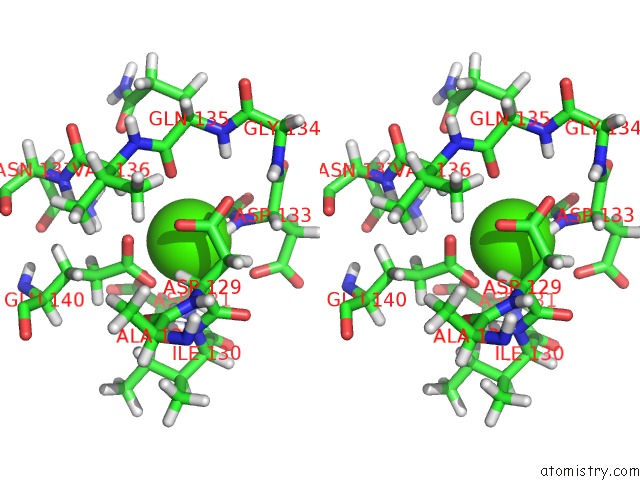
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 4 of Solution uc(Nmr) Structure of Calmodulin/W-7 Complex: the Basis of Diversity in Molecular Recognition, 30 Structures within 5.0Å range:
|
Reference:
M.Osawa,
M.B.Swindells,
J.Tanikawa,
T.Tanaka,
T.Mase,
T.Furuya,
M.Ikura.
Solution Structure of Calmodulin-W-7 Complex: the Basis of Diversity in Molecular Recognition. J.Mol.Biol. V. 276 165 1998.
ISSN: ISSN 0022-2836
PubMed: 9514729
DOI: 10.1006/JMBI.1997.1524
Page generated: Mon Jul 7 17:22:29 2025
ISSN: ISSN 0022-2836
PubMed: 9514729
DOI: 10.1006/JMBI.1997.1524
Last articles
K in 8FHUK in 8F1C
K in 8F1D
K in 8F0N
K in 8F5S
K in 8F5J
K in 8F11
K in 8EYW
K in 8EYV
K in 8F03