Calcium »
PDB 1qd6-1qlk »
1qlk »
Calcium in PDB 1qlk: Solution Structure of Ca(2+)-Loaded Rat S100B (Betabeta) uc(Nmr), 20 Structures
Calcium Binding Sites:
The binding sites of Calcium atom in the Solution Structure of Ca(2+)-Loaded Rat S100B (Betabeta) uc(Nmr), 20 Structures
(pdb code 1qlk). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 4 binding sites of Calcium where determined in the Solution Structure of Ca(2+)-Loaded Rat S100B (Betabeta) uc(Nmr), 20 Structures, PDB code: 1qlk:
Jump to Calcium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Calcium where determined in the Solution Structure of Ca(2+)-Loaded Rat S100B (Betabeta) uc(Nmr), 20 Structures, PDB code: 1qlk:
Jump to Calcium binding site number: 1; 2; 3; 4;
Calcium binding site 1 out of 4 in 1qlk
Go back to
Calcium binding site 1 out
of 4 in the Solution Structure of Ca(2+)-Loaded Rat S100B (Betabeta) uc(Nmr), 20 Structures
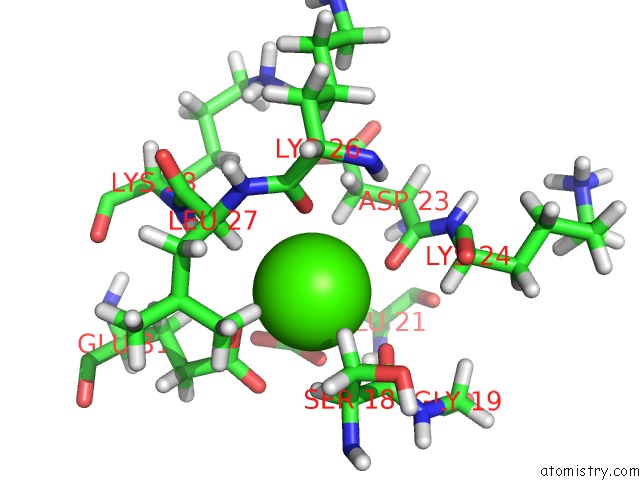
Mono view
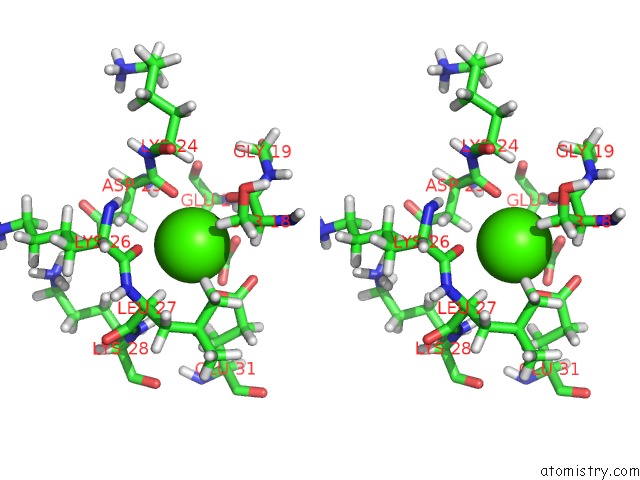
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Solution Structure of Ca(2+)-Loaded Rat S100B (Betabeta) uc(Nmr), 20 Structures within 5.0Å range:
|
Calcium binding site 2 out of 4 in 1qlk
Go back to
Calcium binding site 2 out
of 4 in the Solution Structure of Ca(2+)-Loaded Rat S100B (Betabeta) uc(Nmr), 20 Structures
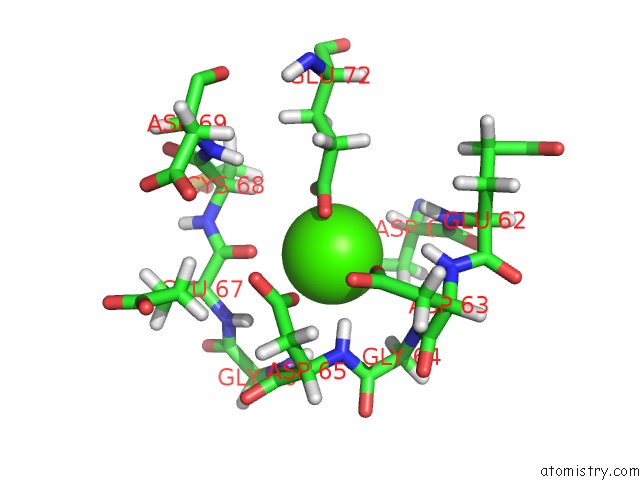
Mono view
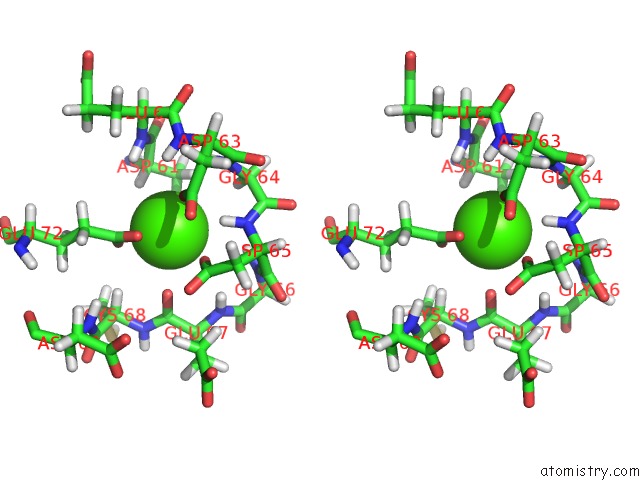
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Solution Structure of Ca(2+)-Loaded Rat S100B (Betabeta) uc(Nmr), 20 Structures within 5.0Å range:
|
Calcium binding site 3 out of 4 in 1qlk
Go back to
Calcium binding site 3 out
of 4 in the Solution Structure of Ca(2+)-Loaded Rat S100B (Betabeta) uc(Nmr), 20 Structures
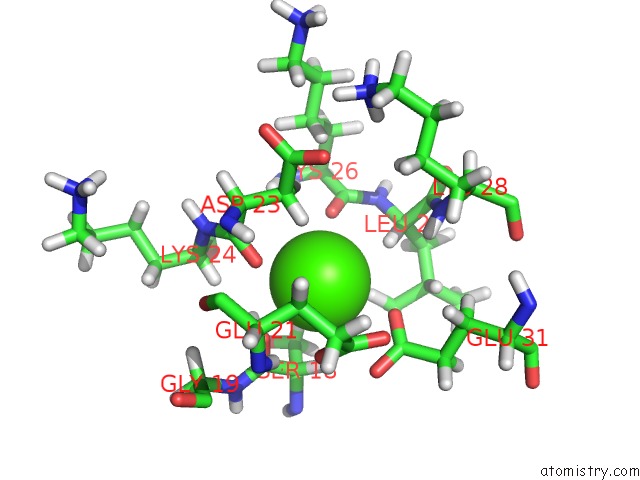
Mono view
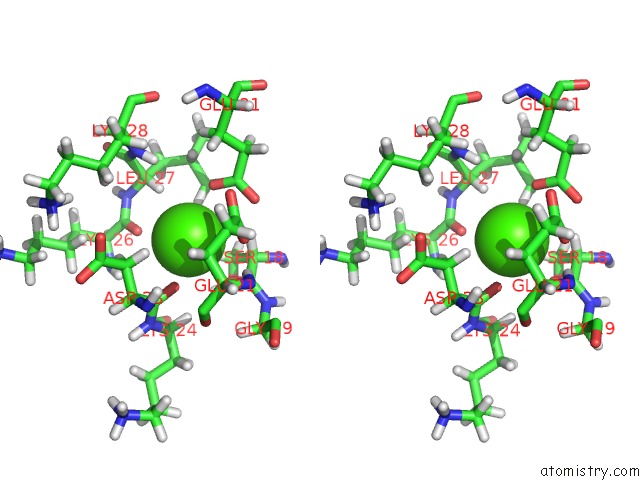
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 3 of Solution Structure of Ca(2+)-Loaded Rat S100B (Betabeta) uc(Nmr), 20 Structures within 5.0Å range:
|
Calcium binding site 4 out of 4 in 1qlk
Go back to
Calcium binding site 4 out
of 4 in the Solution Structure of Ca(2+)-Loaded Rat S100B (Betabeta) uc(Nmr), 20 Structures
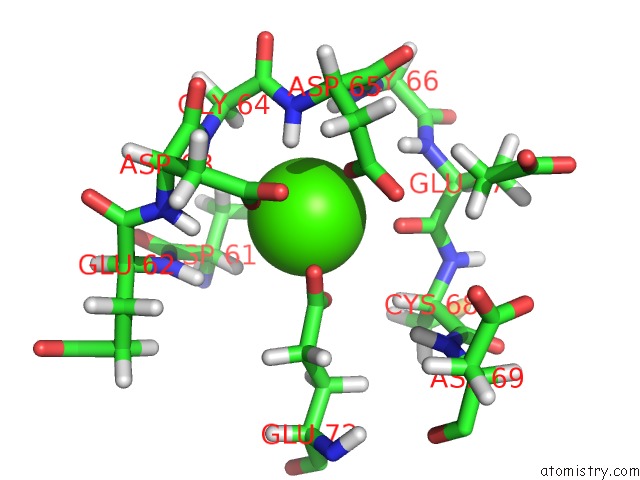
Mono view
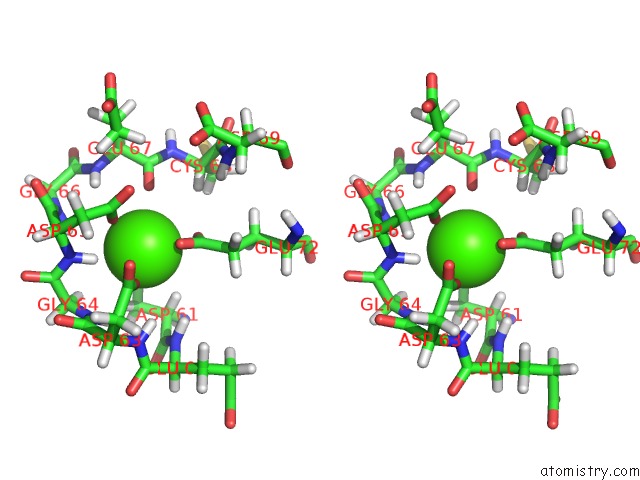
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 4 of Solution Structure of Ca(2+)-Loaded Rat S100B (Betabeta) uc(Nmr), 20 Structures within 5.0Å range:
|
Reference:
A.C.Drohat,
D.M.Baldisseri,
R.R.Rustandi,
D.J.Weber.
Solution Structure of Calcium-Bound Rat S100B(Betabeta) As Determined By Nuclear Magnetic Resonance Spectroscopy,. Biochemistry V. 37 2729 1998.
ISSN: ISSN 0006-2960
PubMed: 9485423
DOI: 10.1021/BI972635P
Page generated: Tue Jul 8 01:24:19 2025
ISSN: ISSN 0006-2960
PubMed: 9485423
DOI: 10.1021/BI972635P
Last articles
Mg in 1VQ4Mg in 1VPA
Mg in 1VPE
Mg in 1VOM
Mg in 1VMA
Mg in 1VMK
Mg in 1VM9
Mg in 1VCR
Mg in 1VLB
Mg in 1VKP