Calcium »
PDB 2jq6-2ksp »
2jul »
Calcium in PDB 2jul: uc(Nmr) Structure of Dream
Calcium Binding Sites:
The binding sites of Calcium atom in the uc(Nmr) Structure of Dream
(pdb code 2jul). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 2 binding sites of Calcium where determined in the uc(Nmr) Structure of Dream, PDB code: 2jul:
Jump to Calcium binding site number: 1; 2;
In total 2 binding sites of Calcium where determined in the uc(Nmr) Structure of Dream, PDB code: 2jul:
Jump to Calcium binding site number: 1; 2;
Calcium binding site 1 out of 2 in 2jul
Go back to
Calcium binding site 1 out
of 2 in the uc(Nmr) Structure of Dream
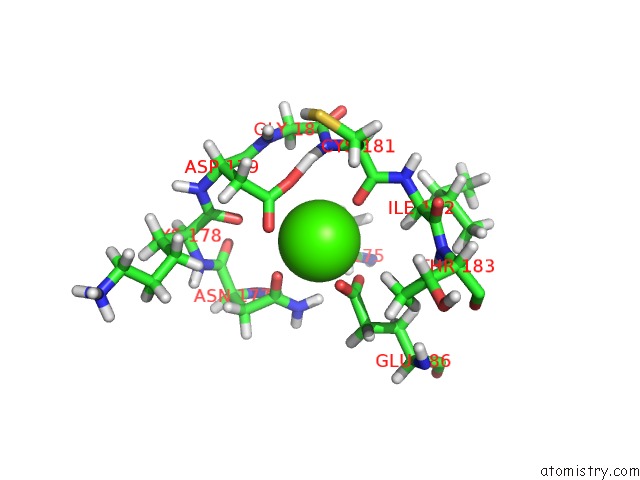
Mono view
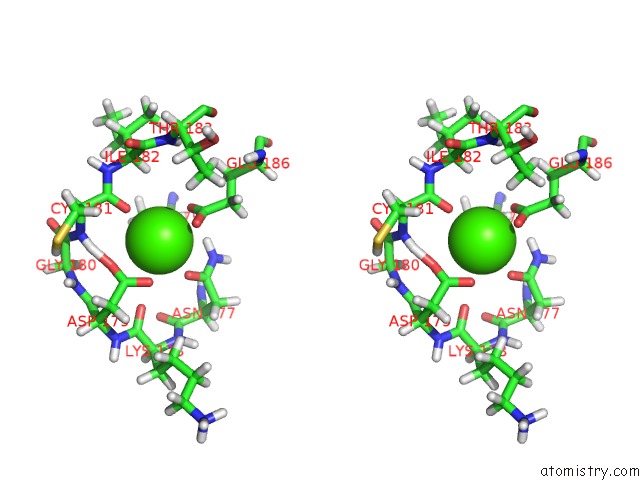
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of uc(Nmr) Structure of Dream within 5.0Å range:
|
Calcium binding site 2 out of 2 in 2jul
Go back to
Calcium binding site 2 out
of 2 in the uc(Nmr) Structure of Dream
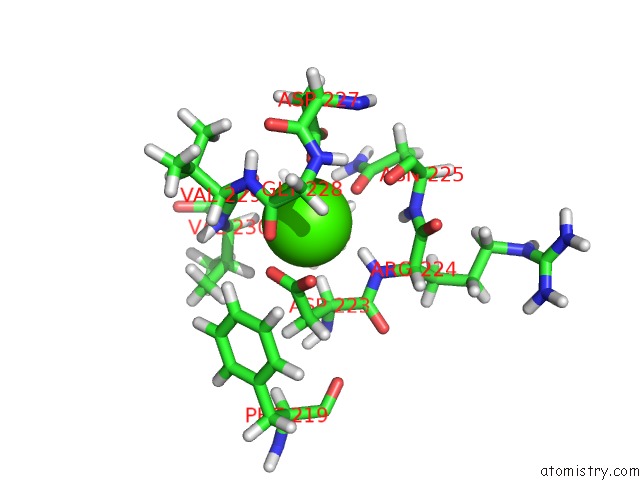
Mono view
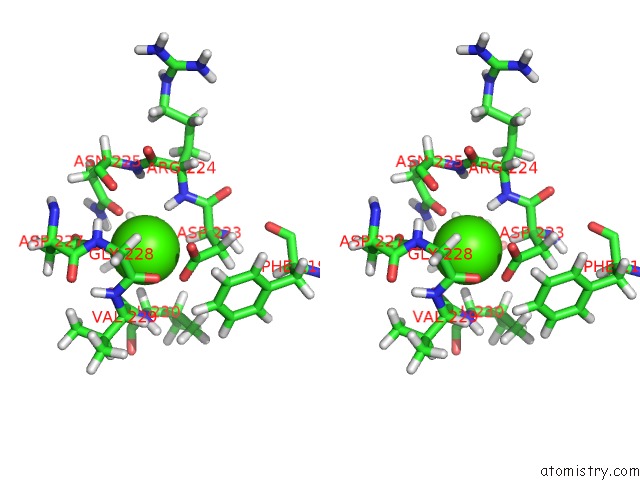
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of uc(Nmr) Structure of Dream within 5.0Å range:
|
Reference:
J.D.Lusin,
M.Vanarotti,
C.Li,
A.Valiveti,
J.B.Ames.
uc(Nmr) Structure of Dream: Implications For Ca(2+)-Dependent Dna Binding and Protein Dimerization. Biochemistry V. 47 2252 2008.
ISSN: ISSN 0006-2960
PubMed: 18201103
DOI: 10.1021/BI7017267
Page generated: Tue Jul 8 06:46:03 2025
ISSN: ISSN 0006-2960
PubMed: 18201103
DOI: 10.1021/BI7017267
Last articles
Mg in 8ZWQMg in 8ZUT
Mg in 8ZVC
Mg in 8ZUS
Mg in 8ZTZ
Mg in 8ZUQ
Mg in 8ZK2
Mg in 8ZUP
Mg in 8ZTF
Mg in 8ZTA