Calcium »
PDB 2m0k-2n8y »
2mzi »
Calcium in PDB 2mzi: uc(Nmr) Solution Structure of the Pro Form of Human Matrilysin (Prommp-7) in Complex with Anionic Membrane
Enzymatic activity of uc(Nmr) Solution Structure of the Pro Form of Human Matrilysin (Prommp-7) in Complex with Anionic Membrane
All present enzymatic activity of uc(Nmr) Solution Structure of the Pro Form of Human Matrilysin (Prommp-7) in Complex with Anionic Membrane:
3.4.24.23;
3.4.24.23;
Other elements in 2mzi:
The structure of uc(Nmr) Solution Structure of the Pro Form of Human Matrilysin (Prommp-7) in Complex with Anionic Membrane also contains other interesting chemical elements:
| Zinc | (Zn) | 40 atoms |
Calcium Binding Sites:
The binding sites of Calcium atom in the uc(Nmr) Solution Structure of the Pro Form of Human Matrilysin (Prommp-7) in Complex with Anionic Membrane
(pdb code 2mzi). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 2 binding sites of Calcium where determined in the uc(Nmr) Solution Structure of the Pro Form of Human Matrilysin (Prommp-7) in Complex with Anionic Membrane, PDB code: 2mzi:
Jump to Calcium binding site number: 1; 2;
In total 2 binding sites of Calcium where determined in the uc(Nmr) Solution Structure of the Pro Form of Human Matrilysin (Prommp-7) in Complex with Anionic Membrane, PDB code: 2mzi:
Jump to Calcium binding site number: 1; 2;
Calcium binding site 1 out of 2 in 2mzi
Go back to
Calcium binding site 1 out
of 2 in the uc(Nmr) Solution Structure of the Pro Form of Human Matrilysin (Prommp-7) in Complex with Anionic Membrane
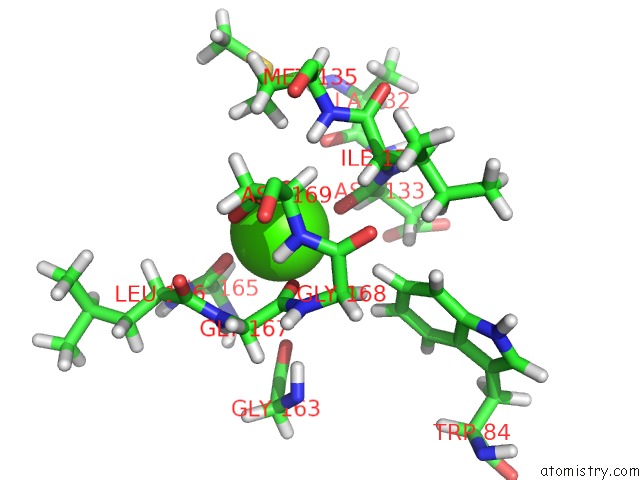
Mono view
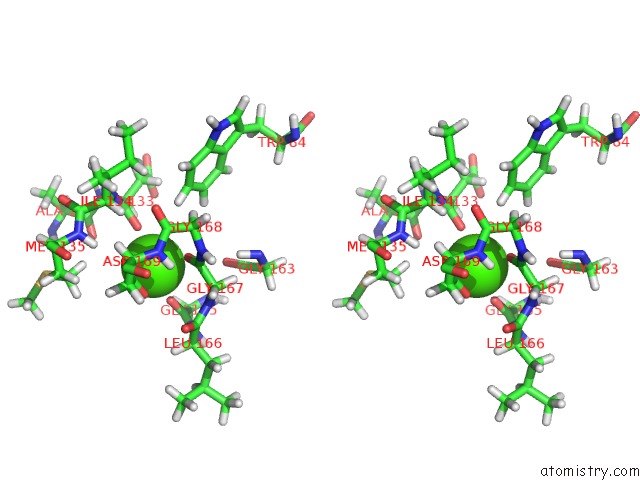
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of uc(Nmr) Solution Structure of the Pro Form of Human Matrilysin (Prommp-7) in Complex with Anionic Membrane within 5.0Å range:
|
Calcium binding site 2 out of 2 in 2mzi
Go back to
Calcium binding site 2 out
of 2 in the uc(Nmr) Solution Structure of the Pro Form of Human Matrilysin (Prommp-7) in Complex with Anionic Membrane
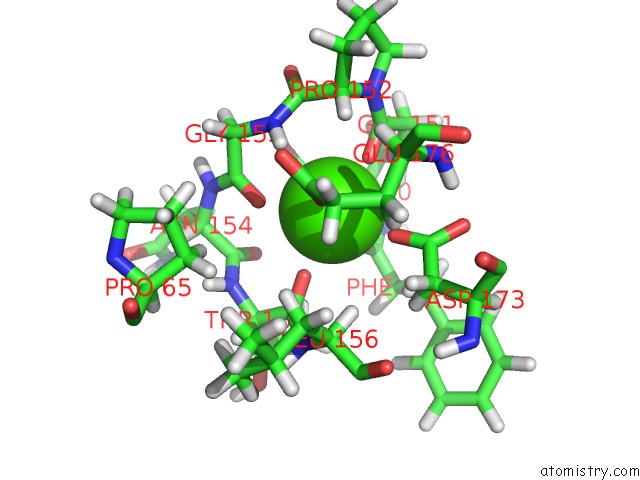
Mono view
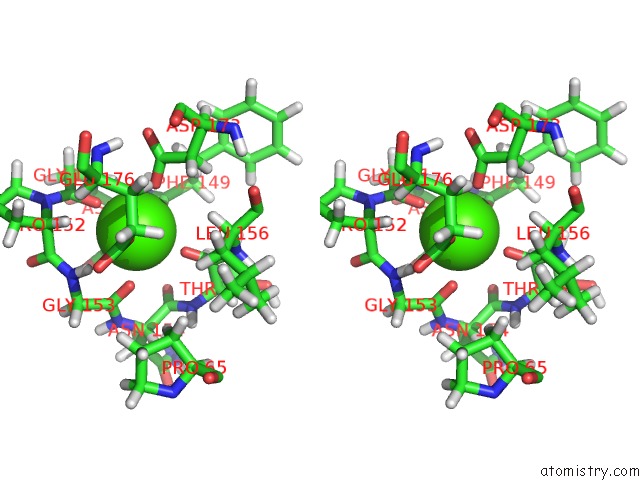
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of uc(Nmr) Solution Structure of the Pro Form of Human Matrilysin (Prommp-7) in Complex with Anionic Membrane within 5.0Å range:
|
Reference:
S.H.Prior,
Y.G.Fulcher,
R.K.Koppisetti,
A.Jurkevich,
S.R.Van Doren.
Charge-Triggered Membrane Insertion of Matrix Metalloproteinase-7, Supporter of Innate Immunity and Tumors. Structure V. 23 2099 2015.
ISSN: ISSN 0969-2126
PubMed: 26439767
DOI: 10.1016/J.STR.2015.08.013
Page generated: Tue Jul 8 07:10:04 2025
ISSN: ISSN 0969-2126
PubMed: 26439767
DOI: 10.1016/J.STR.2015.08.013
Last articles
Mg in 8CGIMg in 8CF8
Mg in 8CGA
Mg in 8CEP
Mg in 8CF1
Mg in 8CAH
Mg in 8CE5
Mg in 8CDQ
Mg in 8CE2
Mg in 8CCO