Calcium »
PDB 2y6j-2yhy »
2ygl »
Calcium in PDB 2ygl: The X-Ray Crystal Structure of Tandem CBM51 Modules of SP3GH98, the Family 98 Glycoside Hydrolase From Streptococcus Pneumoniae SP3-BS71
Protein crystallography data
The structure of The X-Ray Crystal Structure of Tandem CBM51 Modules of SP3GH98, the Family 98 Glycoside Hydrolase From Streptococcus Pneumoniae SP3-BS71, PDB code: 2ygl
was solved by
M.A.Higgins,
E.Ficko-Blean,
C.Wright,
P.J.Meloncelli,
T.L.Lowary,
A.B.Boraston,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 38.91 / 2.10 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 76.040, 90.580, 119.850, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20.953 / 24.671 |
Calcium Binding Sites:
The binding sites of Calcium atom in the The X-Ray Crystal Structure of Tandem CBM51 Modules of SP3GH98, the Family 98 Glycoside Hydrolase From Streptococcus Pneumoniae SP3-BS71
(pdb code 2ygl). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 2 binding sites of Calcium where determined in the The X-Ray Crystal Structure of Tandem CBM51 Modules of SP3GH98, the Family 98 Glycoside Hydrolase From Streptococcus Pneumoniae SP3-BS71, PDB code: 2ygl:
Jump to Calcium binding site number: 1; 2;
In total 2 binding sites of Calcium where determined in the The X-Ray Crystal Structure of Tandem CBM51 Modules of SP3GH98, the Family 98 Glycoside Hydrolase From Streptococcus Pneumoniae SP3-BS71, PDB code: 2ygl:
Jump to Calcium binding site number: 1; 2;
Calcium binding site 1 out of 2 in 2ygl
Go back to
Calcium binding site 1 out
of 2 in the The X-Ray Crystal Structure of Tandem CBM51 Modules of SP3GH98, the Family 98 Glycoside Hydrolase From Streptococcus Pneumoniae SP3-BS71

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of The X-Ray Crystal Structure of Tandem CBM51 Modules of SP3GH98, the Family 98 Glycoside Hydrolase From Streptococcus Pneumoniae SP3-BS71 within 5.0Å range:
|
Calcium binding site 2 out of 2 in 2ygl
Go back to
Calcium binding site 2 out
of 2 in the The X-Ray Crystal Structure of Tandem CBM51 Modules of SP3GH98, the Family 98 Glycoside Hydrolase From Streptococcus Pneumoniae SP3-BS71
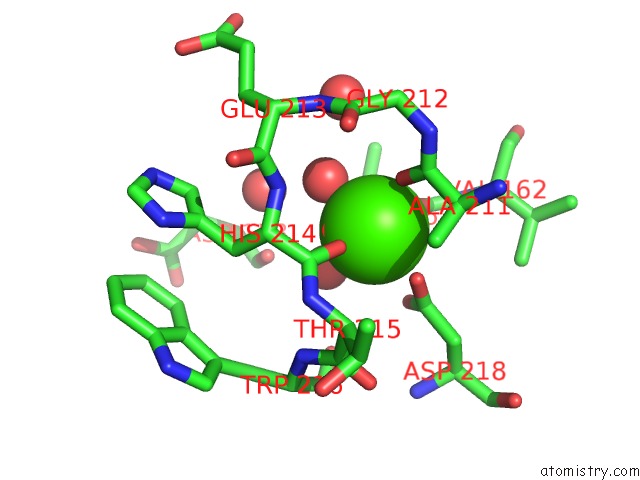
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of The X-Ray Crystal Structure of Tandem CBM51 Modules of SP3GH98, the Family 98 Glycoside Hydrolase From Streptococcus Pneumoniae SP3-BS71 within 5.0Å range:
|
Reference:
M.A.Higgins,
E.Ficko-Blean,
C.Wright,
P.J.Meloncelli,
T.L.Lowary,
A.B.Boraston.
The Overall Architecture and Receptor Binding of Pneumococcal Carbohydrate Antigen Hydrolyzing Enzymes. J.Mol.Biol. V. 411 1017 2011.
ISSN: ISSN 0022-2836
PubMed: 21767550
DOI: 10.1016/J.JMB.2011.06.035
Page generated: Tue Jul 8 09:39:14 2025
ISSN: ISSN 0022-2836
PubMed: 21767550
DOI: 10.1016/J.JMB.2011.06.035
Last articles
F in 8PGNF in 8PGK
F in 8PGJ
F in 8PGI
F in 8PGH
F in 8PGG
F in 8PGE
F in 8PGC
F in 8PGB
F in 8PGA