Calcium »
PDB 3dcq-3dsw »
3de5 »
Calcium in PDB 3de5: Roteinase K By Classical Hanging Drop Method After the Second Step of High X-Ray Dose on Esrf ID23-1 Beamline
Enzymatic activity of Roteinase K By Classical Hanging Drop Method After the Second Step of High X-Ray Dose on Esrf ID23-1 Beamline
All present enzymatic activity of Roteinase K By Classical Hanging Drop Method After the Second Step of High X-Ray Dose on Esrf ID23-1 Beamline:
3.4.21.64;
3.4.21.64;
Protein crystallography data
The structure of Roteinase K By Classical Hanging Drop Method After the Second Step of High X-Ray Dose on Esrf ID23-1 Beamline, PDB code: 3de5
was solved by
E.Pechkova,
S.K.Tripathi,
C.Nicolini,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 35.06 / 2.10 |
| Space group | P 43 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 67.858, 67.858, 102.681, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 16.8 / 23.4 |
Calcium Binding Sites:
The binding sites of Calcium atom in the Roteinase K By Classical Hanging Drop Method After the Second Step of High X-Ray Dose on Esrf ID23-1 Beamline
(pdb code 3de5). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total only one binding site of Calcium was determined in the Roteinase K By Classical Hanging Drop Method After the Second Step of High X-Ray Dose on Esrf ID23-1 Beamline, PDB code: 3de5:
In total only one binding site of Calcium was determined in the Roteinase K By Classical Hanging Drop Method After the Second Step of High X-Ray Dose on Esrf ID23-1 Beamline, PDB code: 3de5:
Calcium binding site 1 out of 1 in 3de5
Go back to
Calcium binding site 1 out
of 1 in the Roteinase K By Classical Hanging Drop Method After the Second Step of High X-Ray Dose on Esrf ID23-1 Beamline
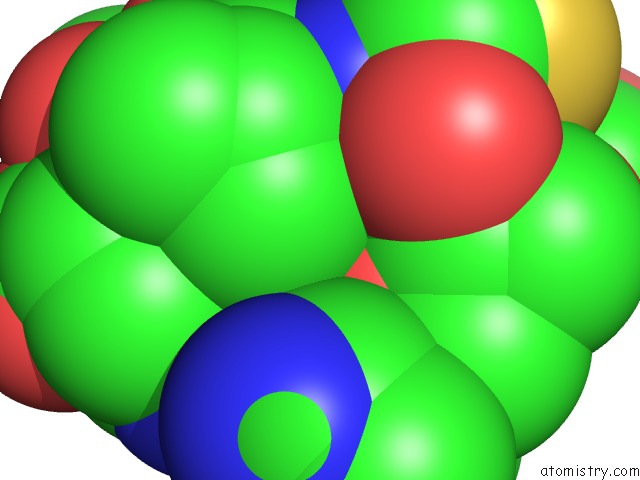
Mono view
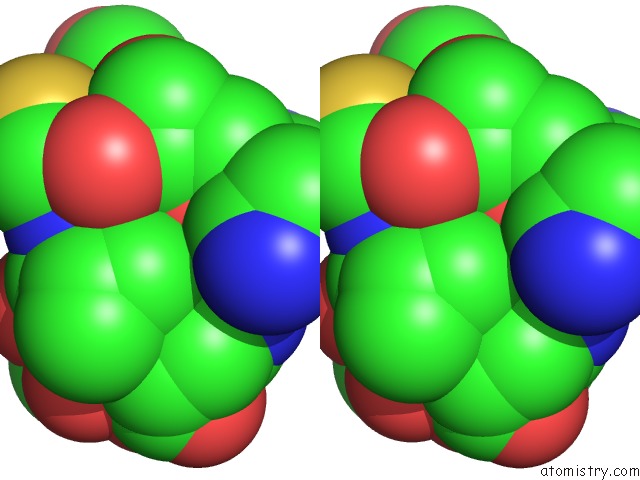
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Roteinase K By Classical Hanging Drop Method After the Second Step of High X-Ray Dose on Esrf ID23-1 Beamline within 5.0Å range:
|
Reference:
E.Pechkova,
S.Tripathi,
R.B.Ravelli,
S.Mcsweeney,
C.Nicolini.
Radiation Stability of Proteinase K Crystals Grown By Lb Nanotemplate Method J.Struct.Biol. V. 168 409 2009.
ISSN: ISSN 1047-8477
PubMed: 19686853
DOI: 10.1016/J.JSB.2009.08.005
Page generated: Tue Jul 8 11:36:18 2025
ISSN: ISSN 1047-8477
PubMed: 19686853
DOI: 10.1016/J.JSB.2009.08.005
Last articles
K in 8CGUK in 8CGR
K in 8CGJ
K in 8CGI
K in 8CG1
K in 8CFY
K in 8CG2
K in 8CFX
K in 8CG0
K in 8CFW