Calcium »
PDB 3qgv-3r3t »
3qlq »
Calcium in PDB 3qlq: Crystal Structure of Concanavalin A Bound to An Octa-Alpha-Mannosyl- Octasilsesquioxane Cluster
Protein crystallography data
The structure of Crystal Structure of Concanavalin A Bound to An Octa-Alpha-Mannosyl- Octasilsesquioxane Cluster, PDB code: 3qlq
was solved by
E.J.Sundberg,
D.A.Bonsor,
B.Trastoy,
J.L.Chiara,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 31.84 / 1.70 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 59.890, 63.680, 125.800, 90.00, 93.23, 90.00 |
| R / Rfree (%) | 21.8 / 24.8 |
Other elements in 3qlq:
The structure of Crystal Structure of Concanavalin A Bound to An Octa-Alpha-Mannosyl- Octasilsesquioxane Cluster also contains other interesting chemical elements:
| Manganese | (Mn) | 4 atoms |
Calcium Binding Sites:
The binding sites of Calcium atom in the Crystal Structure of Concanavalin A Bound to An Octa-Alpha-Mannosyl- Octasilsesquioxane Cluster
(pdb code 3qlq). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 4 binding sites of Calcium where determined in the Crystal Structure of Concanavalin A Bound to An Octa-Alpha-Mannosyl- Octasilsesquioxane Cluster, PDB code: 3qlq:
Jump to Calcium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Calcium where determined in the Crystal Structure of Concanavalin A Bound to An Octa-Alpha-Mannosyl- Octasilsesquioxane Cluster, PDB code: 3qlq:
Jump to Calcium binding site number: 1; 2; 3; 4;
Calcium binding site 1 out of 4 in 3qlq
Go back to
Calcium binding site 1 out
of 4 in the Crystal Structure of Concanavalin A Bound to An Octa-Alpha-Mannosyl- Octasilsesquioxane Cluster
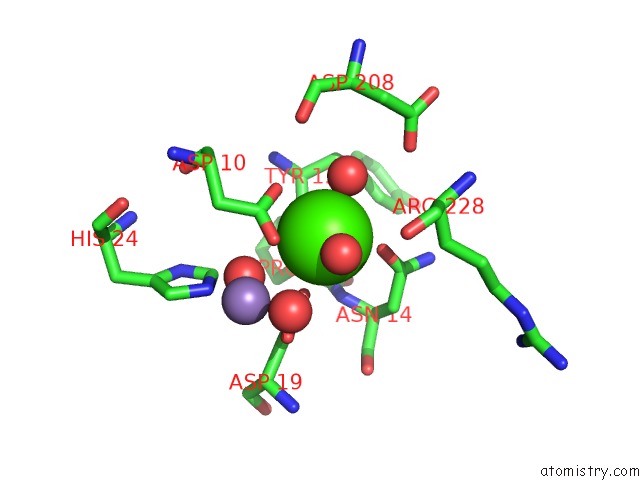
Mono view
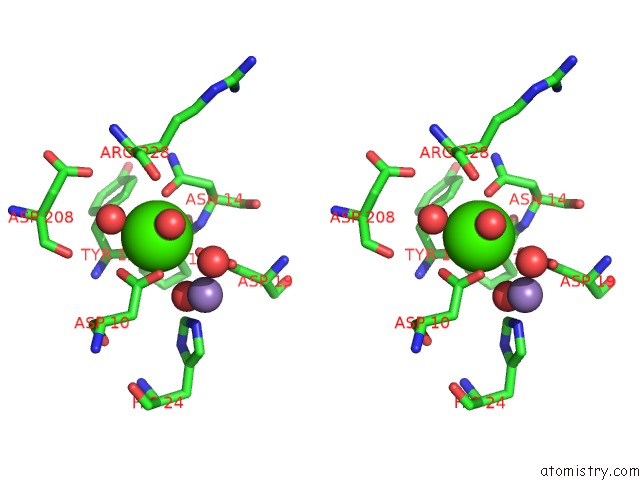
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Crystal Structure of Concanavalin A Bound to An Octa-Alpha-Mannosyl- Octasilsesquioxane Cluster within 5.0Å range:
|
Calcium binding site 2 out of 4 in 3qlq
Go back to
Calcium binding site 2 out
of 4 in the Crystal Structure of Concanavalin A Bound to An Octa-Alpha-Mannosyl- Octasilsesquioxane Cluster
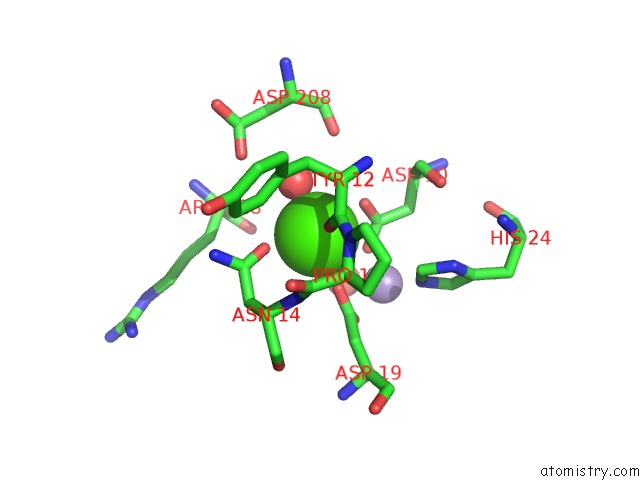
Mono view
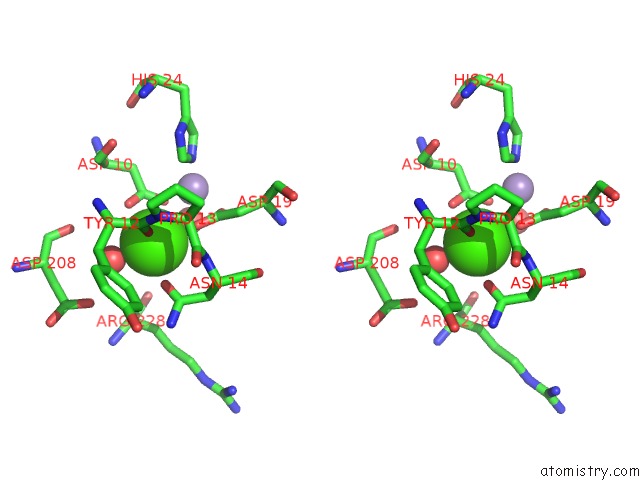
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Crystal Structure of Concanavalin A Bound to An Octa-Alpha-Mannosyl- Octasilsesquioxane Cluster within 5.0Å range:
|
Calcium binding site 3 out of 4 in 3qlq
Go back to
Calcium binding site 3 out
of 4 in the Crystal Structure of Concanavalin A Bound to An Octa-Alpha-Mannosyl- Octasilsesquioxane Cluster
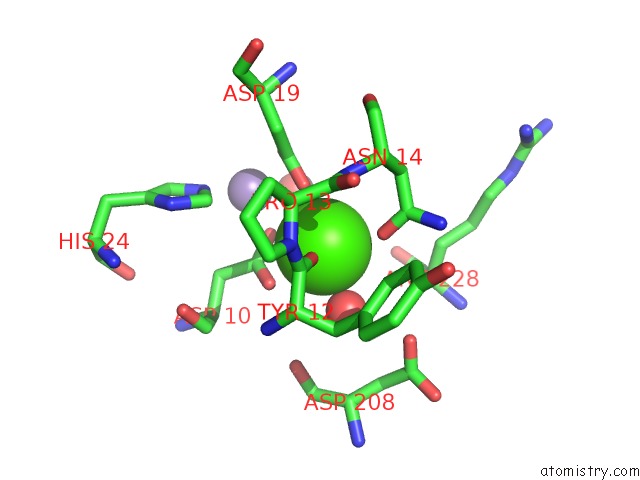
Mono view
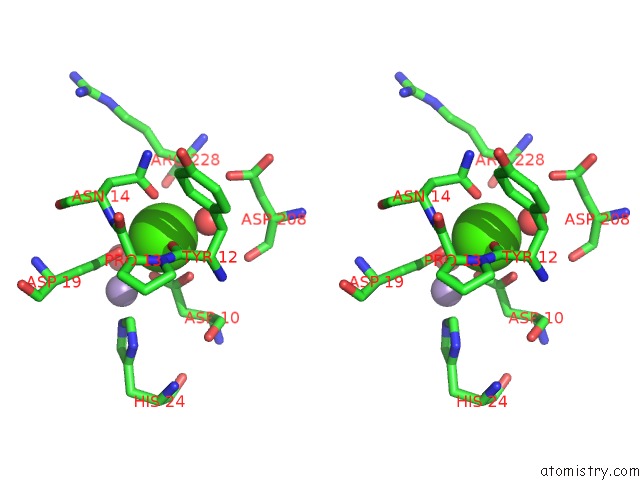
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 3 of Crystal Structure of Concanavalin A Bound to An Octa-Alpha-Mannosyl- Octasilsesquioxane Cluster within 5.0Å range:
|
Calcium binding site 4 out of 4 in 3qlq
Go back to
Calcium binding site 4 out
of 4 in the Crystal Structure of Concanavalin A Bound to An Octa-Alpha-Mannosyl- Octasilsesquioxane Cluster
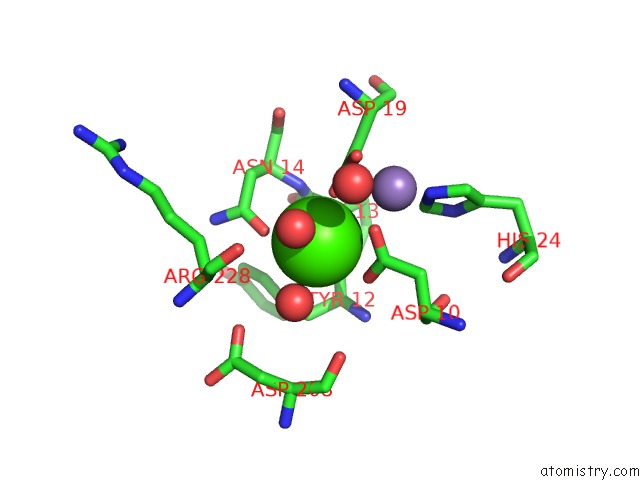
Mono view
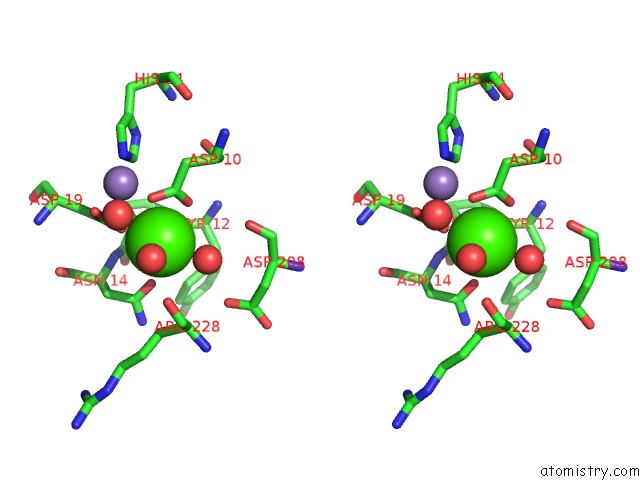
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 4 of Crystal Structure of Concanavalin A Bound to An Octa-Alpha-Mannosyl- Octasilsesquioxane Cluster within 5.0Å range:
|
Reference:
B.Trastoy,
D.A.Bonsor,
M.E.Perez-Ojeda,
E.J.Sundberg,
M.L.Jimeno,
J.M.Garcia-Fernandez,
J.L.Chiara.
Synthesis and Biophysical Study of Disassembling Nano Hybrid Bioconjugates with A Cubic Octasilsesquioxane Core Adv Funct Mater V. 22 3191 2012.
ISSN: ISSN 1616-301X
DOI: 10.1002/ADFM.201200423
Page generated: Tue Jul 8 16:00:45 2025
ISSN: ISSN 1616-301X
DOI: 10.1002/ADFM.201200423
Last articles
K in 2ZXEK in 3A1M
K in 2ZD9
K in 3A3U
K in 2ZWU
K in 2YIA
K in 2ZWT
K in 2ZUJ
K in 2ZUI
K in 2ZUH