Calcium »
PDB 3rke-3rxa »
3rlm »
Calcium in PDB 3rlm: Structure of the W199F Maug/Pre-Methylamine Dehydrogenase Complex After Treatment with Hydrogen Peroxide
Enzymatic activity of Structure of the W199F Maug/Pre-Methylamine Dehydrogenase Complex After Treatment with Hydrogen Peroxide
All present enzymatic activity of Structure of the W199F Maug/Pre-Methylamine Dehydrogenase Complex After Treatment with Hydrogen Peroxide:
1.4.99.3;
1.4.99.3;
Protein crystallography data
The structure of Structure of the W199F Maug/Pre-Methylamine Dehydrogenase Complex After Treatment with Hydrogen Peroxide, PDB code: 3rlm
was solved by
E.T.Yukl,
C.M.Wilmot,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 44.49 / 2.13 |
| Space group | P 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 55.530, 83.520, 107.780, 109.94, 91.54, 105.78 |
| R / Rfree (%) | 18.1 / 23.7 |
Other elements in 3rlm:
The structure of Structure of the W199F Maug/Pre-Methylamine Dehydrogenase Complex After Treatment with Hydrogen Peroxide also contains other interesting chemical elements:
| Iron | (Fe) | 4 atoms |
Calcium Binding Sites:
The binding sites of Calcium atom in the Structure of the W199F Maug/Pre-Methylamine Dehydrogenase Complex After Treatment with Hydrogen Peroxide
(pdb code 3rlm). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 2 binding sites of Calcium where determined in the Structure of the W199F Maug/Pre-Methylamine Dehydrogenase Complex After Treatment with Hydrogen Peroxide, PDB code: 3rlm:
Jump to Calcium binding site number: 1; 2;
In total 2 binding sites of Calcium where determined in the Structure of the W199F Maug/Pre-Methylamine Dehydrogenase Complex After Treatment with Hydrogen Peroxide, PDB code: 3rlm:
Jump to Calcium binding site number: 1; 2;
Calcium binding site 1 out of 2 in 3rlm
Go back to
Calcium binding site 1 out
of 2 in the Structure of the W199F Maug/Pre-Methylamine Dehydrogenase Complex After Treatment with Hydrogen Peroxide
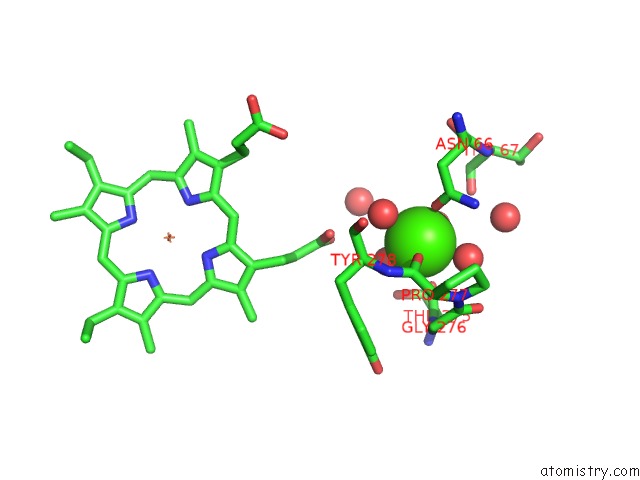
Mono view
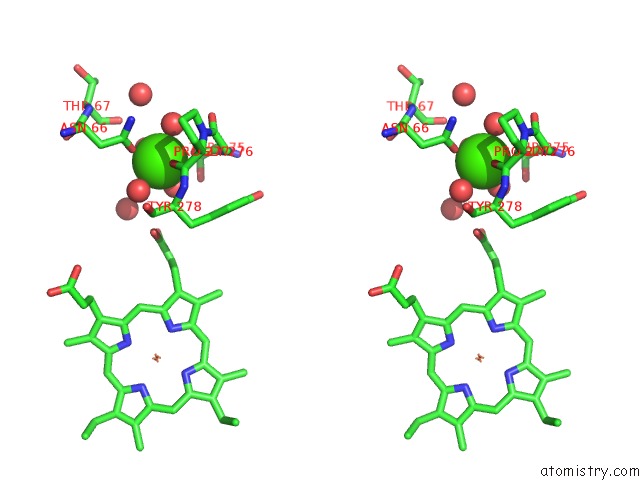
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Structure of the W199F Maug/Pre-Methylamine Dehydrogenase Complex After Treatment with Hydrogen Peroxide within 5.0Å range:
|
Calcium binding site 2 out of 2 in 3rlm
Go back to
Calcium binding site 2 out
of 2 in the Structure of the W199F Maug/Pre-Methylamine Dehydrogenase Complex After Treatment with Hydrogen Peroxide
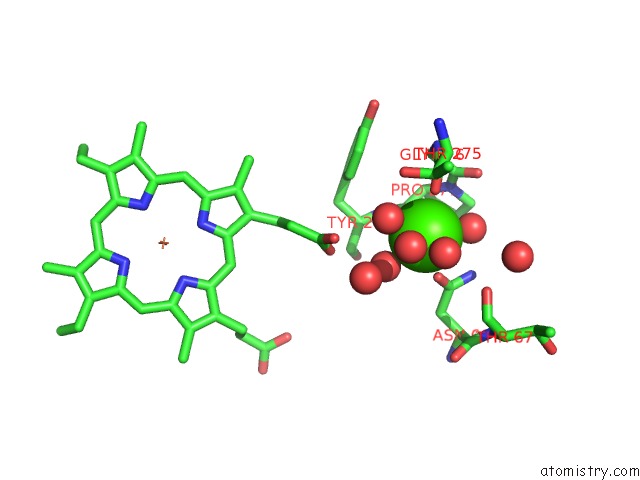
Mono view
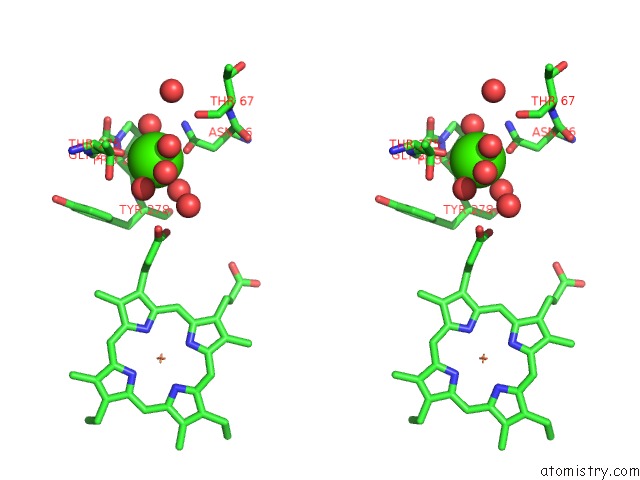
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Structure of the W199F Maug/Pre-Methylamine Dehydrogenase Complex After Treatment with Hydrogen Peroxide within 5.0Å range:
|
Reference:
N.A.Tarboush,
L.M.Jensen,
E.T.Yukl,
J.Geng,
A.Liu,
C.M.Wilmot,
V.L.Davidson.
Mutagenesis of TRYPTOPHAN199 Suggests That Hopping Is Required For Maug-Dependent Tryptophan Tryptophylquinone Biosynthesis. Proc.Natl.Acad.Sci.Usa V. 108 16956 2011.
ISSN: ISSN 0027-8424
PubMed: 21969534
DOI: 10.1073/PNAS.1109423108
Page generated: Tue Jul 8 16:21:17 2025
ISSN: ISSN 0027-8424
PubMed: 21969534
DOI: 10.1073/PNAS.1109423108
Last articles
I in 7C5YI in 7CD0
I in 7CCY
I in 7C3B
I in 7C5W
I in 7C3A
I in 7BFE
I in 7BI2
I in 7BVX
I in 7BHH