Calcium »
PDB 3uix-3v03 »
3usn »
Calcium in PDB 3usn: Structure of the Catalytic Domain of Human Fibroblast Stromelysin-1 Inhibited with the Thiadiazole Inhibitor Ipnu-107859, uc(Nmr), 1 Structure
Enzymatic activity of Structure of the Catalytic Domain of Human Fibroblast Stromelysin-1 Inhibited with the Thiadiazole Inhibitor Ipnu-107859, uc(Nmr), 1 Structure
All present enzymatic activity of Structure of the Catalytic Domain of Human Fibroblast Stromelysin-1 Inhibited with the Thiadiazole Inhibitor Ipnu-107859, uc(Nmr), 1 Structure:
3.4.24.17;
3.4.24.17;
Other elements in 3usn:
The structure of Structure of the Catalytic Domain of Human Fibroblast Stromelysin-1 Inhibited with the Thiadiazole Inhibitor Ipnu-107859, uc(Nmr), 1 Structure also contains other interesting chemical elements:
| Zinc | (Zn) | 2 atoms |
Calcium Binding Sites:
The binding sites of Calcium atom in the Structure of the Catalytic Domain of Human Fibroblast Stromelysin-1 Inhibited with the Thiadiazole Inhibitor Ipnu-107859, uc(Nmr), 1 Structure
(pdb code 3usn). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 3 binding sites of Calcium where determined in the Structure of the Catalytic Domain of Human Fibroblast Stromelysin-1 Inhibited with the Thiadiazole Inhibitor Ipnu-107859, uc(Nmr), 1 Structure, PDB code: 3usn:
Jump to Calcium binding site number: 1; 2; 3;
In total 3 binding sites of Calcium where determined in the Structure of the Catalytic Domain of Human Fibroblast Stromelysin-1 Inhibited with the Thiadiazole Inhibitor Ipnu-107859, uc(Nmr), 1 Structure, PDB code: 3usn:
Jump to Calcium binding site number: 1; 2; 3;
Calcium binding site 1 out of 3 in 3usn
Go back to
Calcium binding site 1 out
of 3 in the Structure of the Catalytic Domain of Human Fibroblast Stromelysin-1 Inhibited with the Thiadiazole Inhibitor Ipnu-107859, uc(Nmr), 1 Structure
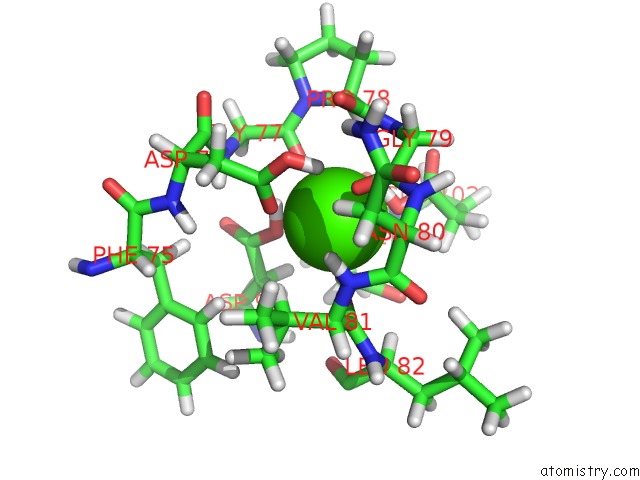
Mono view
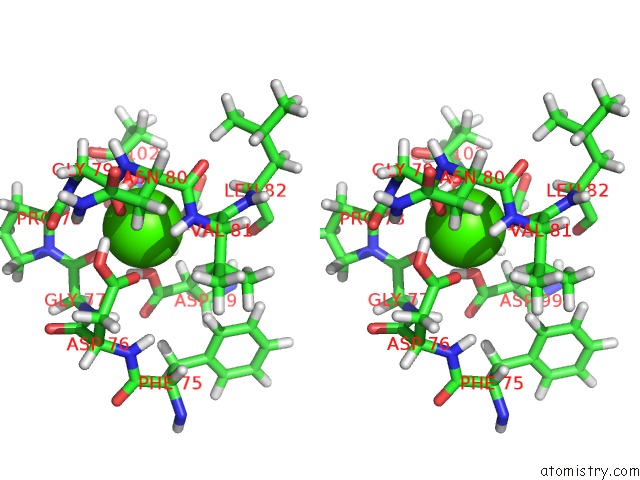
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Structure of the Catalytic Domain of Human Fibroblast Stromelysin-1 Inhibited with the Thiadiazole Inhibitor Ipnu-107859, uc(Nmr), 1 Structure within 5.0Å range:
|
Calcium binding site 2 out of 3 in 3usn
Go back to
Calcium binding site 2 out
of 3 in the Structure of the Catalytic Domain of Human Fibroblast Stromelysin-1 Inhibited with the Thiadiazole Inhibitor Ipnu-107859, uc(Nmr), 1 Structure
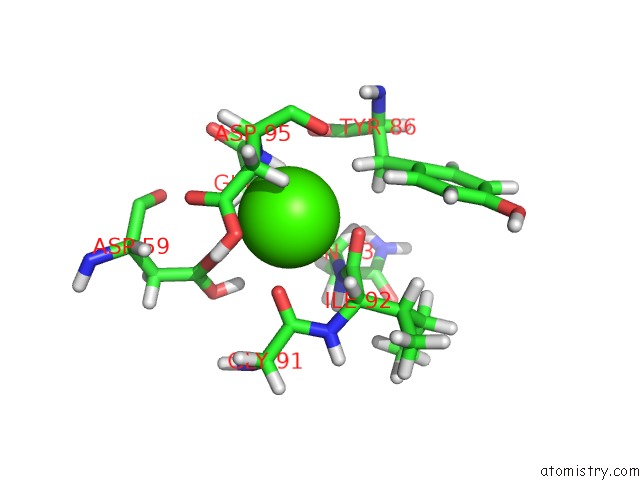
Mono view
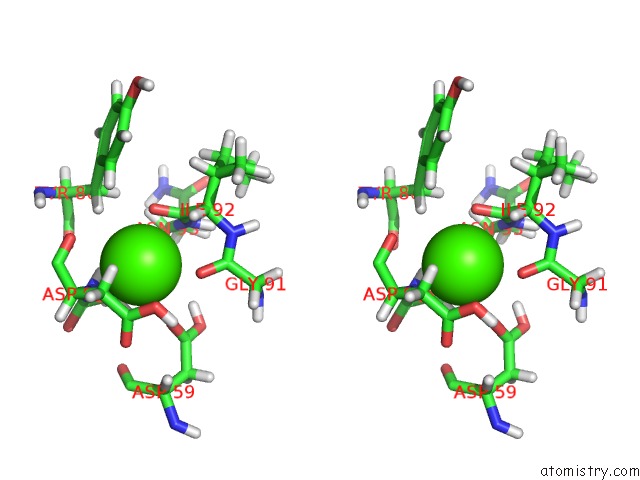
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Structure of the Catalytic Domain of Human Fibroblast Stromelysin-1 Inhibited with the Thiadiazole Inhibitor Ipnu-107859, uc(Nmr), 1 Structure within 5.0Å range:
|
Calcium binding site 3 out of 3 in 3usn
Go back to
Calcium binding site 3 out
of 3 in the Structure of the Catalytic Domain of Human Fibroblast Stromelysin-1 Inhibited with the Thiadiazole Inhibitor Ipnu-107859, uc(Nmr), 1 Structure
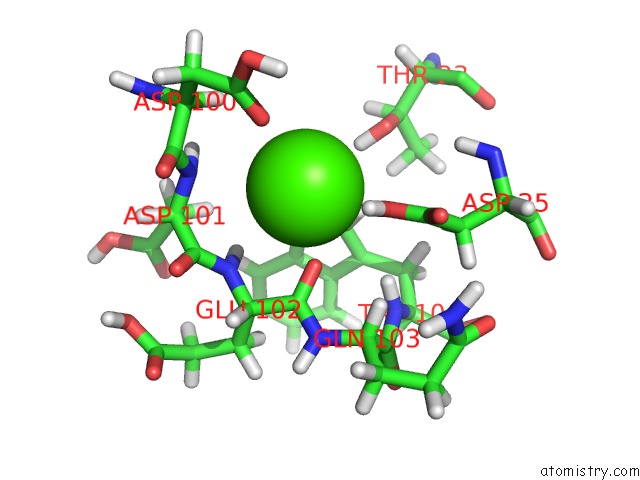
Mono view
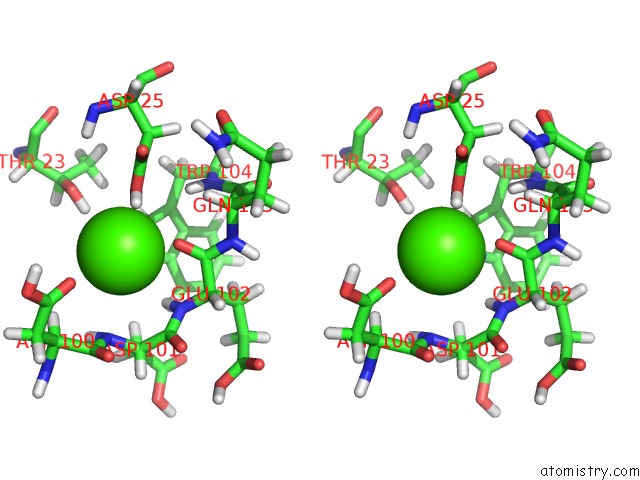
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 3 of Structure of the Catalytic Domain of Human Fibroblast Stromelysin-1 Inhibited with the Thiadiazole Inhibitor Ipnu-107859, uc(Nmr), 1 Structure within 5.0Å range:
|
Reference:
B.J.Stockman,
D.J.Waldon,
J.A.Gates,
T.A.Scahill,
D.A.Kloosterman,
S.A.Mizsak,
E.J.Jacobsen,
K.L.Belonga,
M.A.Mitchell,
B.Mao,
J.D.Petke,
L.Goodman,
E.A.Powers,
S.R.Ledbetter,
P.S.Kaytes,
G.Vogeli,
V.P.Marshall,
G.L.Petzold,
R.A.Poorman.
Solution Structures of Stromelysin Complexed to Thiadiazole Inhibitors. Protein Sci. V. 7 2281 1998.
ISSN: ISSN 0961-8368
PubMed: 9827994
Page generated: Tue Jul 8 17:19:51 2025
ISSN: ISSN 0961-8368
PubMed: 9827994
Last articles
Mg in 6C90Mg in 6CA0
Mg in 6C9Y
Mg in 6C8Z
Mg in 6C8P
Mg in 6C8N
Mg in 6C8O
Mg in 6C8D
Mg in 6C8L
Mg in 6C8J