Calcium »
PDB 4q1o-4qn4 »
4q6q »
Calcium in PDB 4q6q: Structural Analysis of the Zn-Form II of Helicobacter Pylori CSD4, A D,L-Carboxypeptidase
Protein crystallography data
The structure of Structural Analysis of the Zn-Form II of Helicobacter Pylori CSD4, A D,L-Carboxypeptidase, PDB code: 4q6q
was solved by
H.S.Kim,
J.Kim,
H.N.Im,
D.R.An,
M.Lee,
D.Hesek,
S.Mobashery,
J.Y.Kim,
K.Cho,
H.J.Yoon,
B.W.Han,
B.I.Lee,
S.W.Suh,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 30.00 / 2.40 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 53.314, 66.928, 144.806, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 16.7 / 20.5 |
Other elements in 4q6q:
The structure of Structural Analysis of the Zn-Form II of Helicobacter Pylori CSD4, A D,L-Carboxypeptidase also contains other interesting chemical elements:
| Zinc | (Zn) | 2 atoms |
Calcium Binding Sites:
The binding sites of Calcium atom in the Structural Analysis of the Zn-Form II of Helicobacter Pylori CSD4, A D,L-Carboxypeptidase
(pdb code 4q6q). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 2 binding sites of Calcium where determined in the Structural Analysis of the Zn-Form II of Helicobacter Pylori CSD4, A D,L-Carboxypeptidase, PDB code: 4q6q:
Jump to Calcium binding site number: 1; 2;
In total 2 binding sites of Calcium where determined in the Structural Analysis of the Zn-Form II of Helicobacter Pylori CSD4, A D,L-Carboxypeptidase, PDB code: 4q6q:
Jump to Calcium binding site number: 1; 2;
Calcium binding site 1 out of 2 in 4q6q
Go back to
Calcium binding site 1 out
of 2 in the Structural Analysis of the Zn-Form II of Helicobacter Pylori CSD4, A D,L-Carboxypeptidase
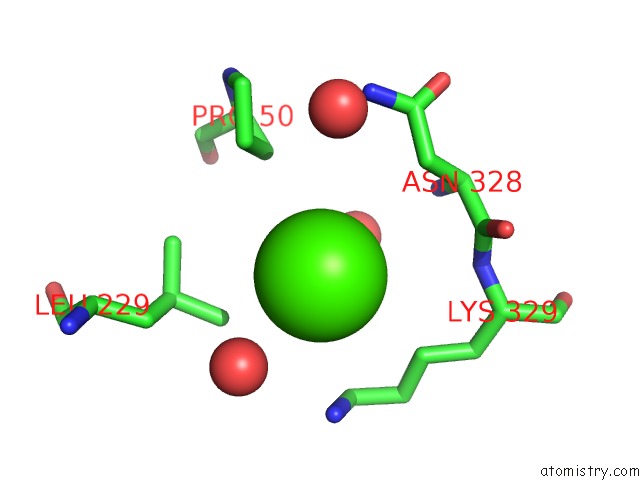
Mono view
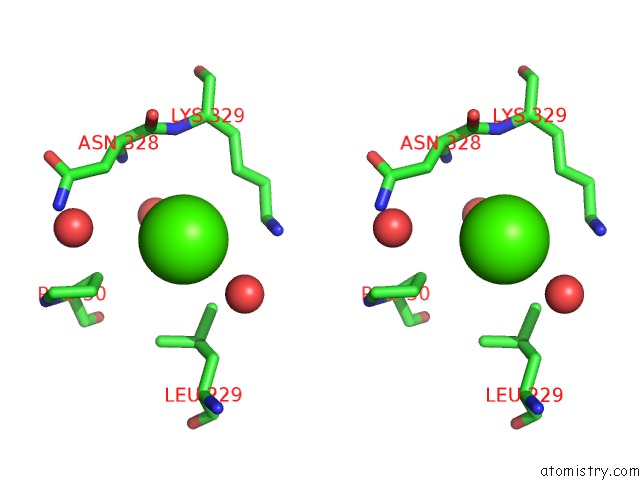
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Structural Analysis of the Zn-Form II of Helicobacter Pylori CSD4, A D,L-Carboxypeptidase within 5.0Å range:
|
Calcium binding site 2 out of 2 in 4q6q
Go back to
Calcium binding site 2 out
of 2 in the Structural Analysis of the Zn-Form II of Helicobacter Pylori CSD4, A D,L-Carboxypeptidase
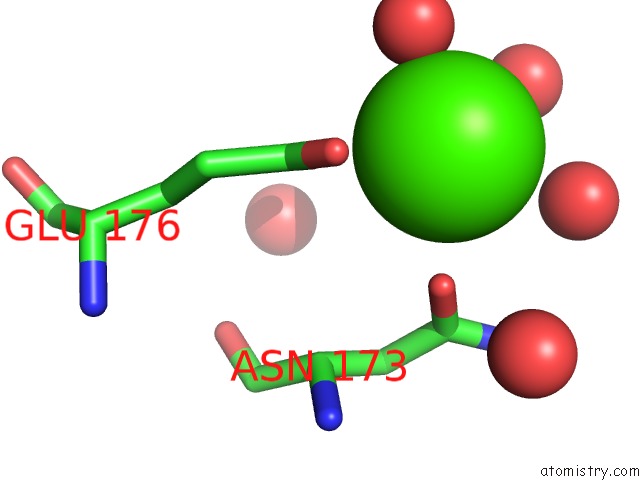
Mono view
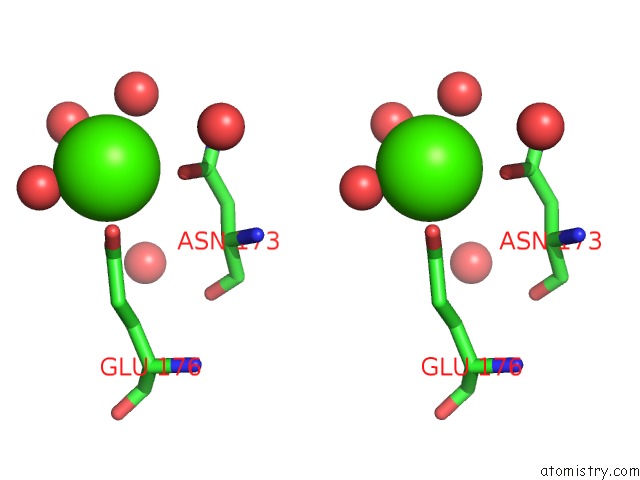
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Structural Analysis of the Zn-Form II of Helicobacter Pylori CSD4, A D,L-Carboxypeptidase within 5.0Å range:
|
Reference:
H.S.Kim,
J.Kim,
H.N.Im,
D.R.An,
M.Lee,
D.Hesek,
S.Mobashery,
J.Y.Kim,
K.Cho,
H.J.Yoon,
B.W.Han,
B.I.Lee,
S.W.Suh.
Structural Basis For the Recognition of Muramyltripeptide By Helicobacter Pylori CSD4, A D,L-Carboxypeptidase Controlling the Helical Cell Shape Acta Crystallogr.,Sect.D V. 70 2014.
ISSN: ESSN 1399-0047
DOI: 10.1107/S1399004714018732
Page generated: Wed Jul 9 01:41:31 2025
ISSN: ESSN 1399-0047
DOI: 10.1107/S1399004714018732
Last articles
Mg in 4GMJMg in 4GNK
Mg in 4GNI
Mg in 4GN0
Mg in 4GMX
Mg in 4GME
Mg in 4GKM
Mg in 4GKR
Mg in 4GHL
Mg in 4GIU