Calcium »
PDB 5jap-5jrc »
5jee »
Calcium in PDB 5jee: Apo-Structure of Humanised Rada-Mutant HUMRADA26F
Protein crystallography data
The structure of Apo-Structure of Humanised Rada-Mutant HUMRADA26F, PDB code: 5jee
was solved by
G.Fischer,
M.Marsh,
T.Moschetti,
T.Sharpe,
D.Scott,
M.Morgan,
H.Ng,
J.Skidmore,
A.Venkitaraman,
C.Abell,
T.L.Blundell,
M.Hyvonen,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 37.13 / 1.49 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 50.581, 50.597, 74.269, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 19.4 / 21.8 |
Calcium Binding Sites:
The binding sites of Calcium atom in the Apo-Structure of Humanised Rada-Mutant HUMRADA26F
(pdb code 5jee). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total only one binding site of Calcium was determined in the Apo-Structure of Humanised Rada-Mutant HUMRADA26F, PDB code: 5jee:
In total only one binding site of Calcium was determined in the Apo-Structure of Humanised Rada-Mutant HUMRADA26F, PDB code: 5jee:
Calcium binding site 1 out of 1 in 5jee
Go back to
Calcium binding site 1 out
of 1 in the Apo-Structure of Humanised Rada-Mutant HUMRADA26F
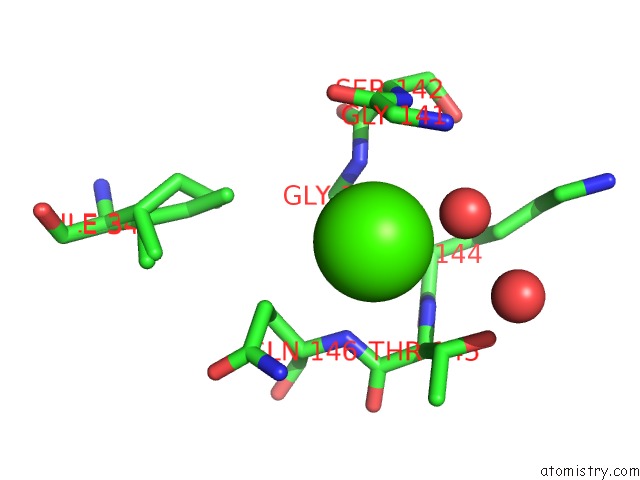
Mono view
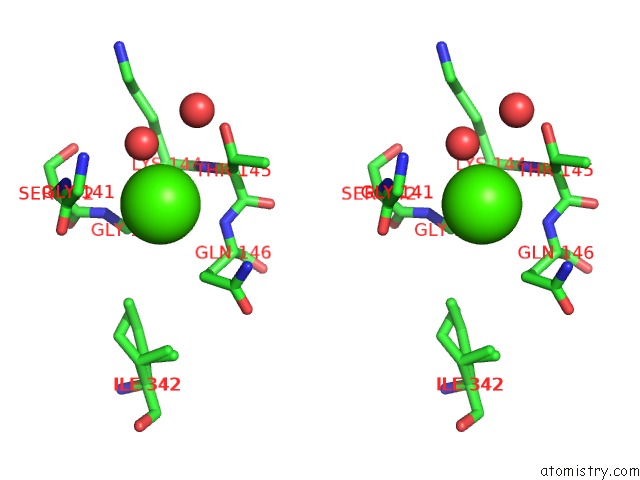
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Apo-Structure of Humanised Rada-Mutant HUMRADA26F within 5.0Å range:
|
Reference:
T.Moschetti,
T.Sharpe,
G.Fischer,
M.E.Marsh,
H.K.Ng,
M.Morgan,
D.E.Scott,
T.L.Blundell,
A.R Venkitaraman,
J.Skidmore,
C.Abell,
M.Hyvonen.
Engineering Archeal Surrogate Systems For the Development of Protein-Protein Interaction Inhibitors Against Human RAD51. J.Mol.Biol. V. 428 4589 2016.
ISSN: ESSN 1089-8638
PubMed: 27725183
DOI: 10.1016/J.JMB.2016.10.009
Page generated: Wed Jul 9 07:07:13 2025
ISSN: ESSN 1089-8638
PubMed: 27725183
DOI: 10.1016/J.JMB.2016.10.009
Last articles
K in 3HRZK in 3HPL
K in 3H8G
K in 3HQN
K in 3H8F
K in 3HHE
K in 3HOG
K in 3HJW
K in 3HFW
K in 3HC1