Calcium »
PDB 6oly-6p8x »
6p48 »
Calcium in PDB 6p48: Cryo-Em Structure of Calcium-Bound TMEM16F in Nanodisc with Supplement of PIP2 in CL1
Calcium Binding Sites:
The binding sites of Calcium atom in the Cryo-Em Structure of Calcium-Bound TMEM16F in Nanodisc with Supplement of PIP2 in CL1
(pdb code 6p48). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 2 binding sites of Calcium where determined in the Cryo-Em Structure of Calcium-Bound TMEM16F in Nanodisc with Supplement of PIP2 in CL1, PDB code: 6p48:
Jump to Calcium binding site number: 1; 2;
In total 2 binding sites of Calcium where determined in the Cryo-Em Structure of Calcium-Bound TMEM16F in Nanodisc with Supplement of PIP2 in CL1, PDB code: 6p48:
Jump to Calcium binding site number: 1; 2;
Calcium binding site 1 out of 2 in 6p48
Go back to
Calcium binding site 1 out
of 2 in the Cryo-Em Structure of Calcium-Bound TMEM16F in Nanodisc with Supplement of PIP2 in CL1
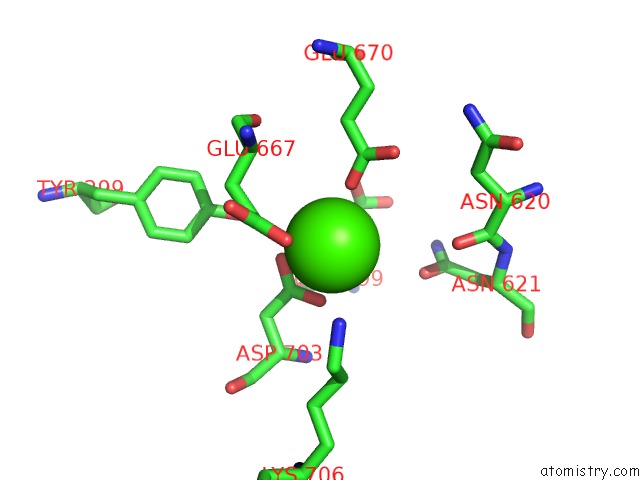
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Cryo-Em Structure of Calcium-Bound TMEM16F in Nanodisc with Supplement of PIP2 in CL1 within 5.0Å range:
|
Calcium binding site 2 out of 2 in 6p48
Go back to
Calcium binding site 2 out
of 2 in the Cryo-Em Structure of Calcium-Bound TMEM16F in Nanodisc with Supplement of PIP2 in CL1
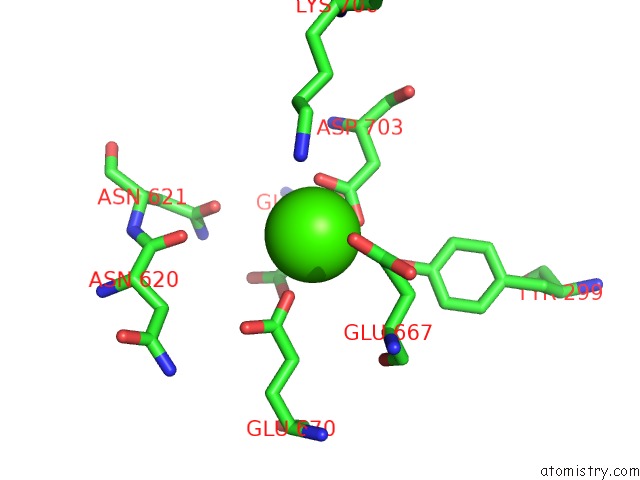
Mono view
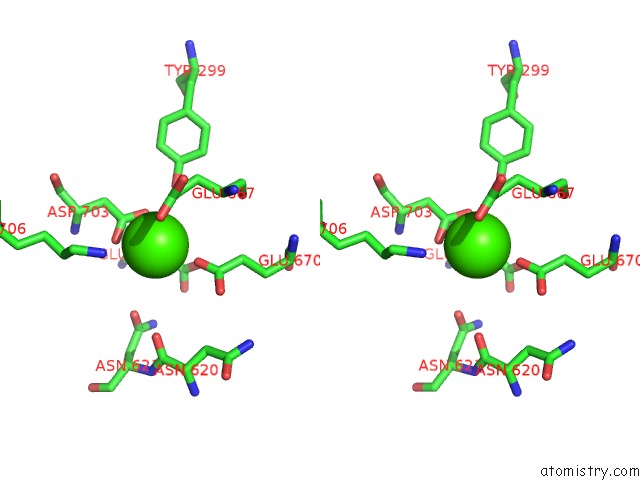
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Cryo-Em Structure of Calcium-Bound TMEM16F in Nanodisc with Supplement of PIP2 in CL1 within 5.0Å range:
|
Reference:
S.Feng,
S.Dang,
T.W.Han,
W.Ye,
P.Jin,
T.Cheng,
J.Li,
Y.N.Jan,
L.Y.Jan,
Y.Cheng.
Cryo-Em Studies of TMEM16F Calcium-Activated Ion Channel Suggest Features Important For Lipid Scrambling. Cell Rep V. 28 567 2019.
ISSN: ESSN 2211-1247
PubMed: 31291589
DOI: 10.1016/J.CELREP.2019.06.023
Page generated: Wed Jul 9 16:42:59 2025
ISSN: ESSN 2211-1247
PubMed: 31291589
DOI: 10.1016/J.CELREP.2019.06.023
Last articles
Mg in 4GMJMg in 4GNK
Mg in 4GNI
Mg in 4GN0
Mg in 4GMX
Mg in 4GME
Mg in 4GKM
Mg in 4GKR
Mg in 4GHL
Mg in 4GIU