Calcium »
PDB 7dlh-7e6q »
7dxi »
Calcium in PDB 7dxi: Structure of Drosophila Melanogaster Glcnac-1-Phosphotransferase
Calcium Binding Sites:
The binding sites of Calcium atom in the Structure of Drosophila Melanogaster Glcnac-1-Phosphotransferase
(pdb code 7dxi). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 2 binding sites of Calcium where determined in the Structure of Drosophila Melanogaster Glcnac-1-Phosphotransferase, PDB code: 7dxi:
Jump to Calcium binding site number: 1; 2;
In total 2 binding sites of Calcium where determined in the Structure of Drosophila Melanogaster Glcnac-1-Phosphotransferase, PDB code: 7dxi:
Jump to Calcium binding site number: 1; 2;
Calcium binding site 1 out of 2 in 7dxi
Go back to
Calcium binding site 1 out
of 2 in the Structure of Drosophila Melanogaster Glcnac-1-Phosphotransferase
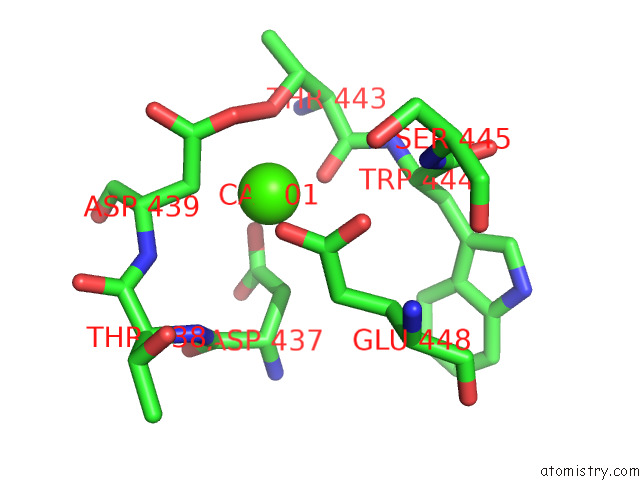
Mono view
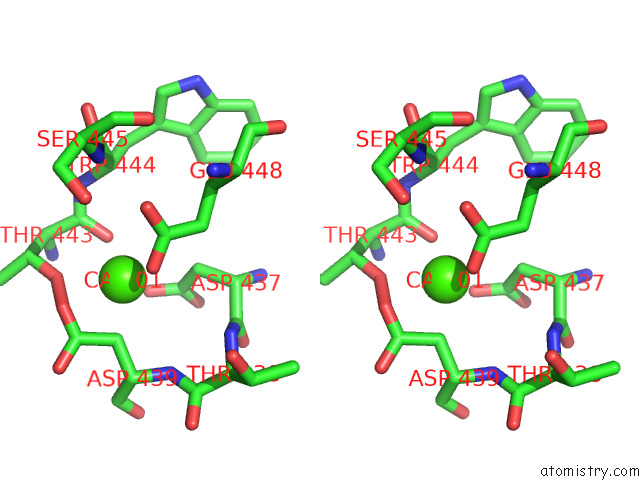
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Structure of Drosophila Melanogaster Glcnac-1-Phosphotransferase within 5.0Å range:
|
Calcium binding site 2 out of 2 in 7dxi
Go back to
Calcium binding site 2 out
of 2 in the Structure of Drosophila Melanogaster Glcnac-1-Phosphotransferase
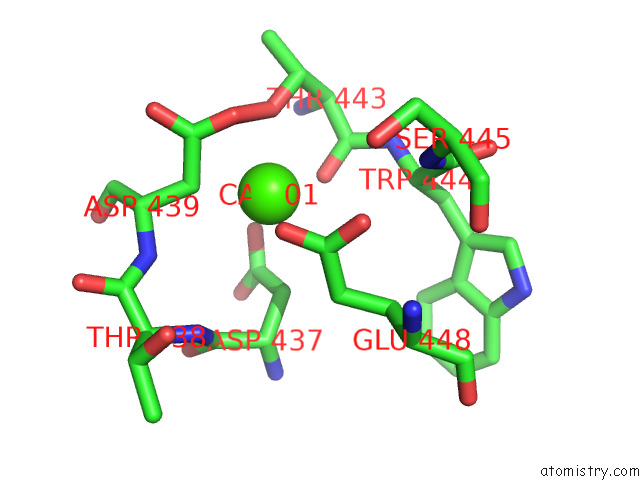
Mono view
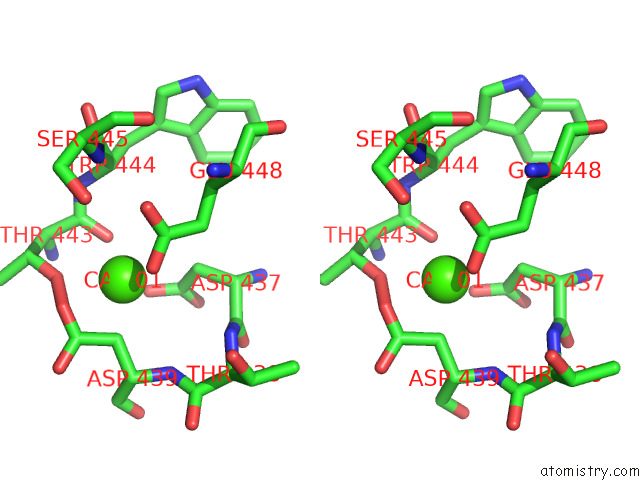
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Structure of Drosophila Melanogaster Glcnac-1-Phosphotransferase within 5.0Å range:
|
Reference:
S.Du,
G.Wang,
Z.Zhang,
C.Ma,
N.Gao,
J.Xiao.
Structural Insights Into How Glcnac-1-Phosphotransferase Directs Lysosomal Protein Transport. J.Biol.Chem. V. 298 01702 2022.
ISSN: ESSN 1083-351X
PubMed: 35148990
DOI: 10.1016/J.JBC.2022.101702
Page generated: Wed Jul 9 21:42:21 2025
ISSN: ESSN 1083-351X
PubMed: 35148990
DOI: 10.1016/J.JBC.2022.101702
Last articles
Mg in 4DUXMg in 4DUW
Mg in 4DUV
Mg in 4DUO
Mg in 4DUG
Mg in 4DTY
Mg in 4DTW
Mg in 4DTH
Mg in 4DTF
Mg in 4DSU