Calcium »
PDB 7pod-7qfy »
7qfi »
Calcium in PDB 7qfi: Crystal Structure of S-Layer Protein Slpx From Lactobacillus Acidophilus, Domain I (Aa 31-182)
Protein crystallography data
The structure of Crystal Structure of S-Layer Protein Slpx From Lactobacillus Acidophilus, Domain I (Aa 31-182), PDB code: 7qfi
was solved by
T.Sagmeister,
E.Damisch,
C.Millan,
I.Uson,
M.Eder,
T.Pavkov-Keller,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 41.28 / 1.50 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 37.35, 38.347, 82.745, 90, 93.93, 90 |
| R / Rfree (%) | 18.7 / 22.2 |
Calcium Binding Sites:
The binding sites of Calcium atom in the Crystal Structure of S-Layer Protein Slpx From Lactobacillus Acidophilus, Domain I (Aa 31-182)
(pdb code 7qfi). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total only one binding site of Calcium was determined in the Crystal Structure of S-Layer Protein Slpx From Lactobacillus Acidophilus, Domain I (Aa 31-182), PDB code: 7qfi:
In total only one binding site of Calcium was determined in the Crystal Structure of S-Layer Protein Slpx From Lactobacillus Acidophilus, Domain I (Aa 31-182), PDB code: 7qfi:
Calcium binding site 1 out of 1 in 7qfi
Go back to
Calcium binding site 1 out
of 1 in the Crystal Structure of S-Layer Protein Slpx From Lactobacillus Acidophilus, Domain I (Aa 31-182)
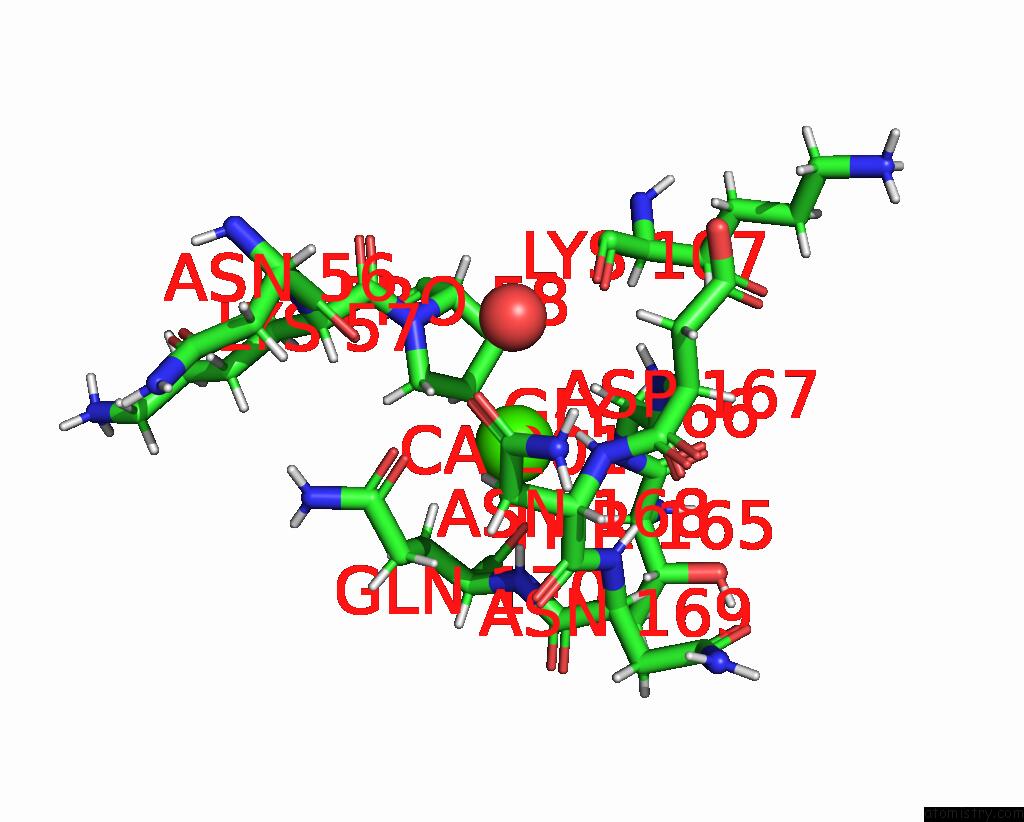
Mono view
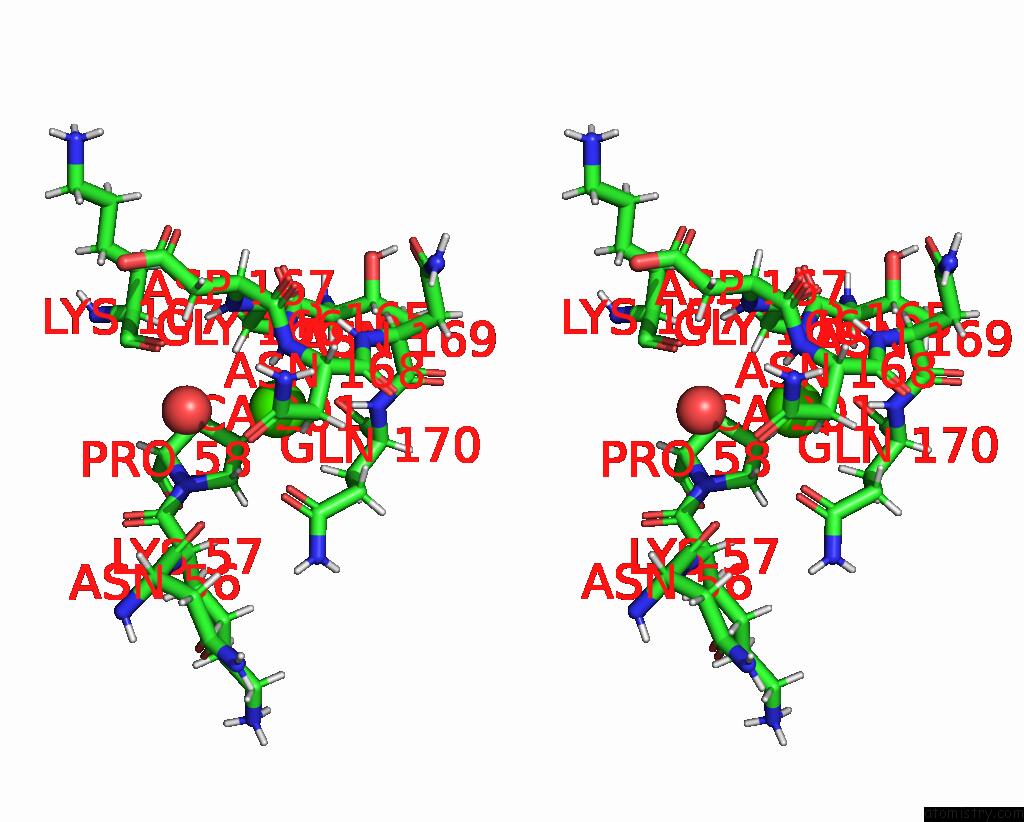
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Crystal Structure of S-Layer Protein Slpx From Lactobacillus Acidophilus, Domain I (Aa 31-182) within 5.0Å range:
|
Reference:
T.Sagmeister,
M.Eder,
C.Grininger,
D.Vejzovic,
C.Buhlheller,
E.Damisch,
C.Millan,
A.Medina,
I.Uson,
M.Baek,
R.Read,
D.Baker,
T.Pavkov-Keller.
The Self-Assembly of the S-Layer Protein From Lactobacilli Acidophilus To Be Published.
Page generated: Thu Jul 10 00:34:17 2025
Last articles
K in 6AFZK in 6AFY
K in 6AFX
K in 6AFW
K in 6AFV
K in 6AFU
K in 6AFT
K in 6AFS
K in 6ABM
K in 6AB2