Calcium »
PDB 9bbf-9cm9 »
9cic »
Calcium in PDB 9cic: Solution uc(Nmr) Structure of Calcium-Bound I89, A Dynamic Engineered Protein
Calcium Binding Sites:
The binding sites of Calcium atom in the Solution uc(Nmr) Structure of Calcium-Bound I89, A Dynamic Engineered Protein
(pdb code 9cic). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total only one binding site of Calcium was determined in the Solution uc(Nmr) Structure of Calcium-Bound I89, A Dynamic Engineered Protein, PDB code: 9cic:
In total only one binding site of Calcium was determined in the Solution uc(Nmr) Structure of Calcium-Bound I89, A Dynamic Engineered Protein, PDB code: 9cic:
Calcium binding site 1 out of 1 in 9cic
Go back to
Calcium binding site 1 out
of 1 in the Solution uc(Nmr) Structure of Calcium-Bound I89, A Dynamic Engineered Protein
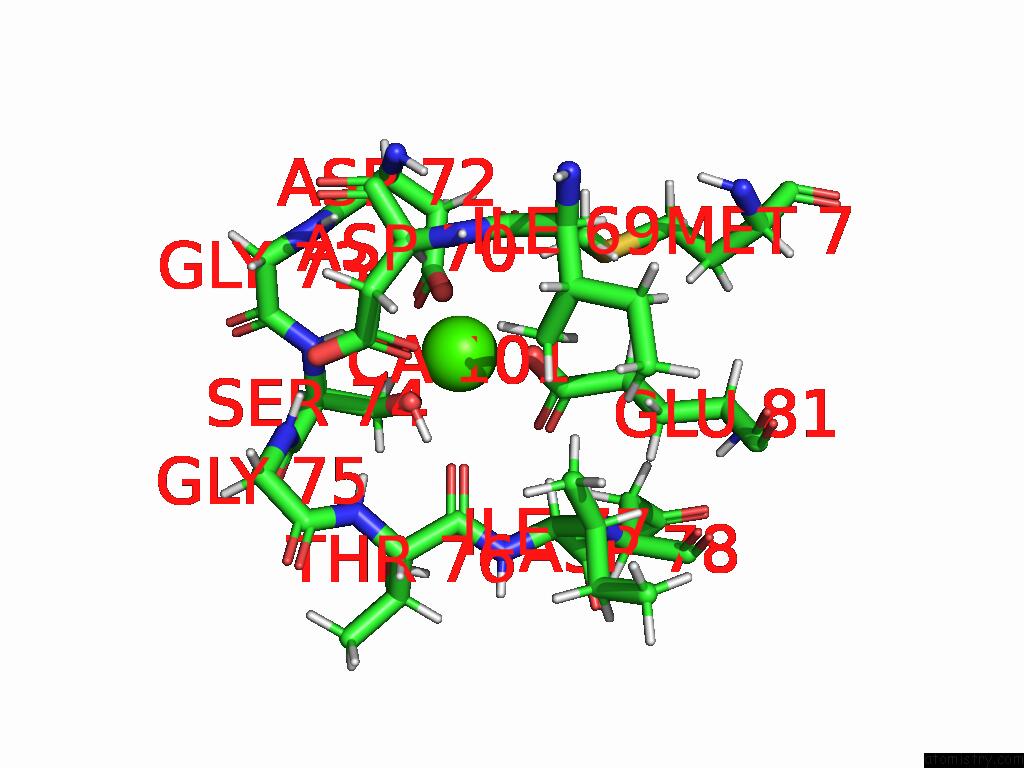
Mono view
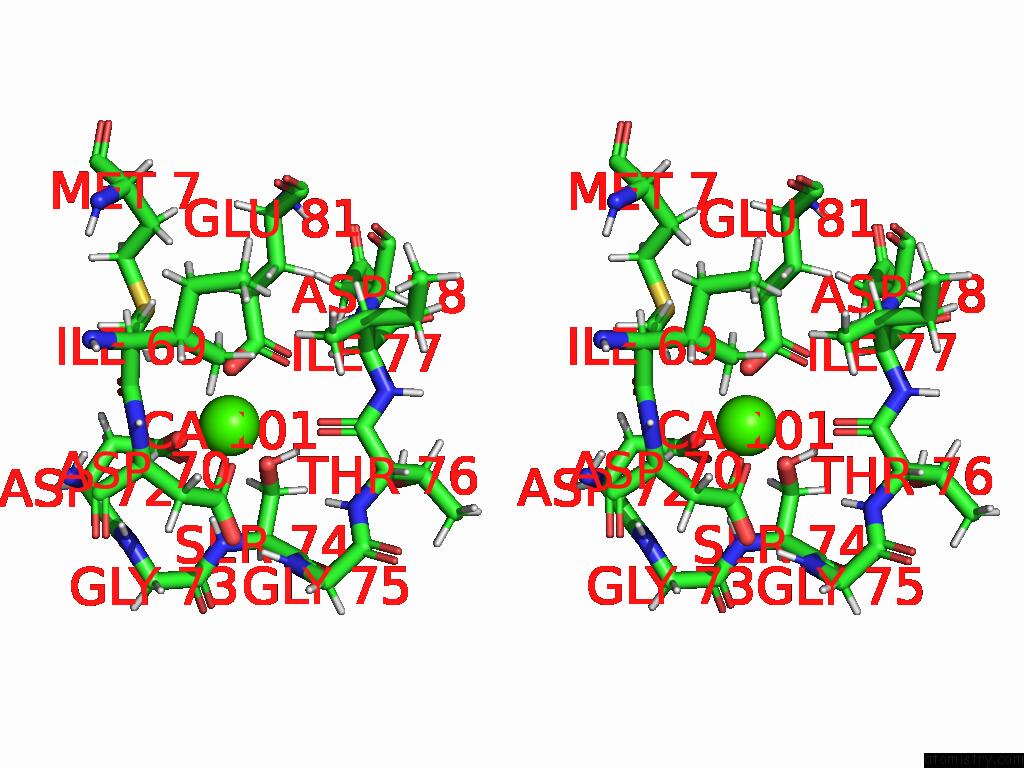
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Solution uc(Nmr) Structure of Calcium-Bound I89, A Dynamic Engineered Protein within 5.0Å range:
|
Reference:
A.B.Guo,
D.Akpinaroglu,
C.A.Stephens,
M.Grabe,
C.A.Smith,
M.J.S.Kelly,
T.Kortemme.
Deep Learning-Guided Design of Dynamic Proteins Science 2025.
ISSN: ESSN 1095-9203
DOI: 10.1126/SCIENCE.ADR7094
Page generated: Thu Jul 10 09:04:41 2025
ISSN: ESSN 1095-9203
DOI: 10.1126/SCIENCE.ADR7094
Last articles
Mn in 9LJUMn in 9LJW
Mn in 9LJS
Mn in 9LJR
Mn in 9LJT
Mn in 9LJV
Mg in 9UA2
Mg in 9R96
Mg in 9VM1
Mg in 9P01