Calcium »
PDB 9cp0-9dqt »
9d5a »
Calcium in PDB 9d5a: Structure of Citrobacter Multi-Ubiquitin Protein, Local Refinement of One Full-Length Protomer
Calcium Binding Sites:
The binding sites of Calcium atom in the Structure of Citrobacter Multi-Ubiquitin Protein, Local Refinement of One Full-Length Protomer
(pdb code 9d5a). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 5 binding sites of Calcium where determined in the Structure of Citrobacter Multi-Ubiquitin Protein, Local Refinement of One Full-Length Protomer, PDB code: 9d5a:
Jump to Calcium binding site number: 1; 2; 3; 4; 5;
In total 5 binding sites of Calcium where determined in the Structure of Citrobacter Multi-Ubiquitin Protein, Local Refinement of One Full-Length Protomer, PDB code: 9d5a:
Jump to Calcium binding site number: 1; 2; 3; 4; 5;
Calcium binding site 1 out of 5 in 9d5a
Go back to
Calcium binding site 1 out
of 5 in the Structure of Citrobacter Multi-Ubiquitin Protein, Local Refinement of One Full-Length Protomer
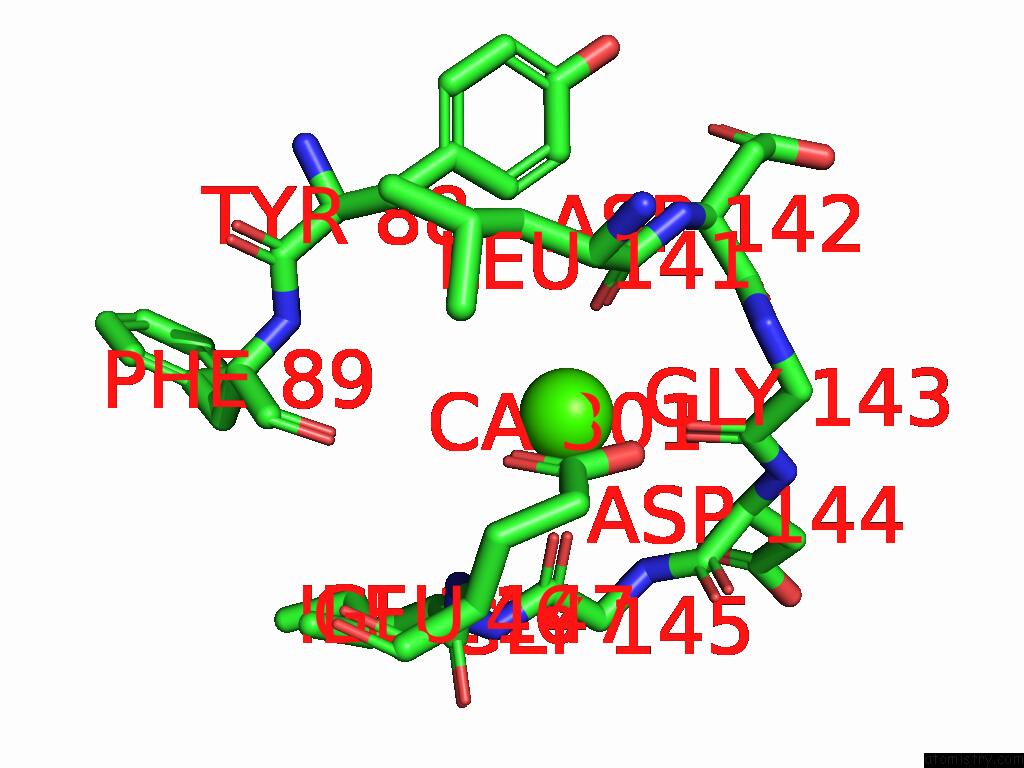
Mono view
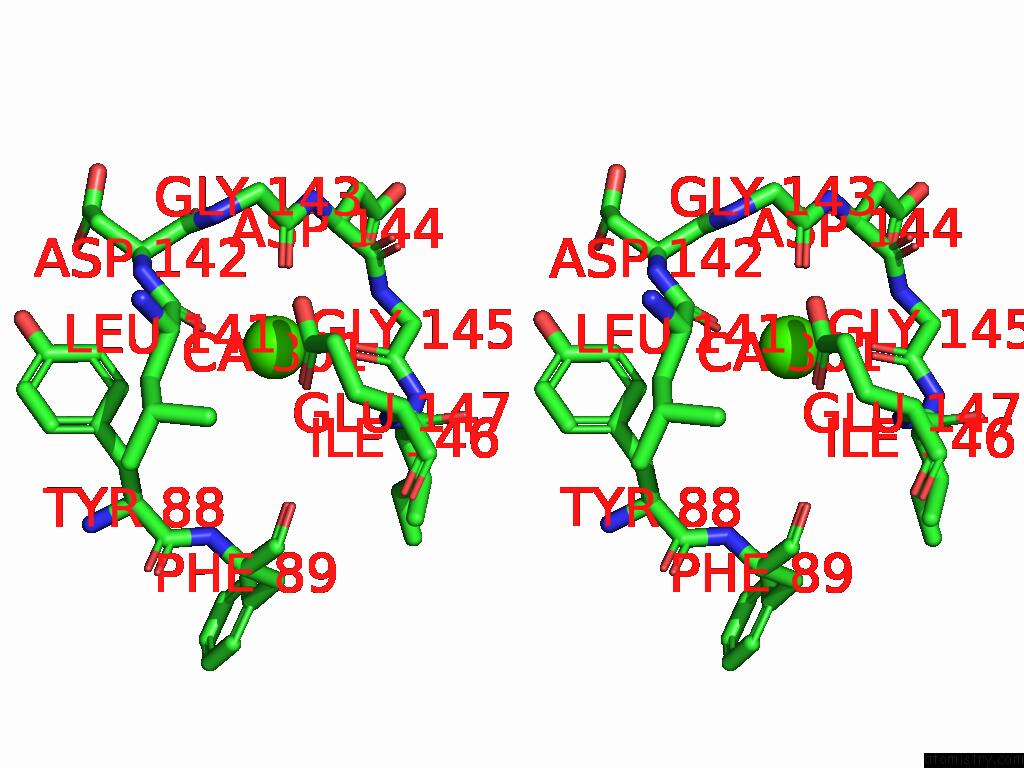
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Structure of Citrobacter Multi-Ubiquitin Protein, Local Refinement of One Full-Length Protomer within 5.0Å range:
|
Calcium binding site 2 out of 5 in 9d5a
Go back to
Calcium binding site 2 out
of 5 in the Structure of Citrobacter Multi-Ubiquitin Protein, Local Refinement of One Full-Length Protomer
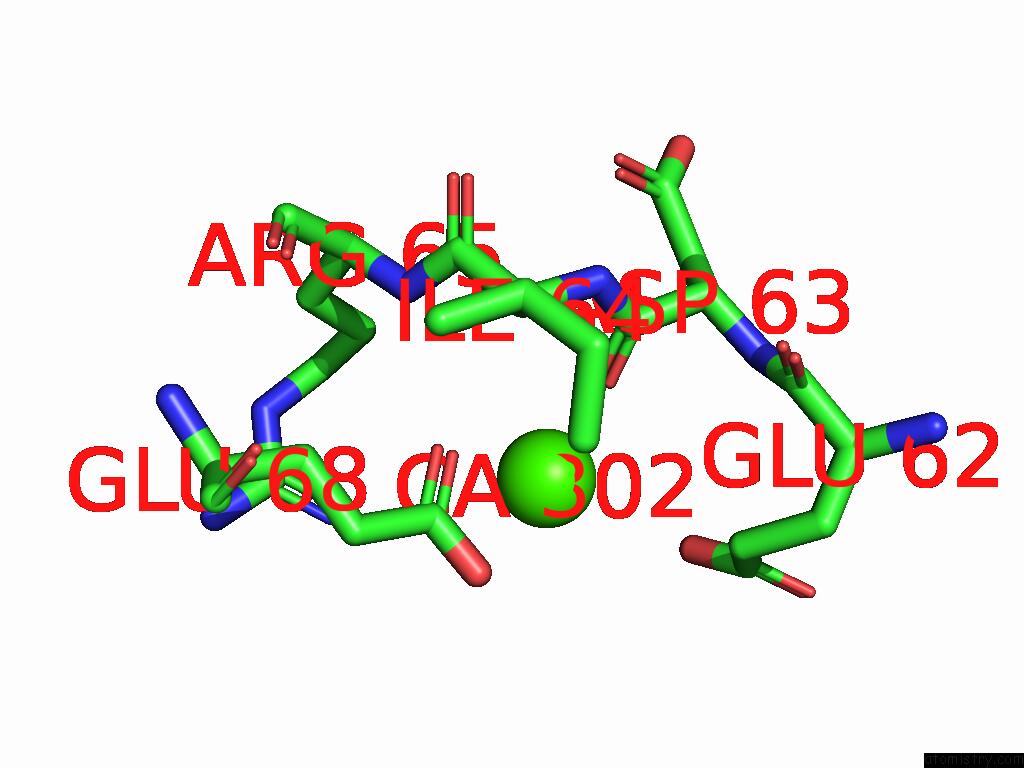
Mono view
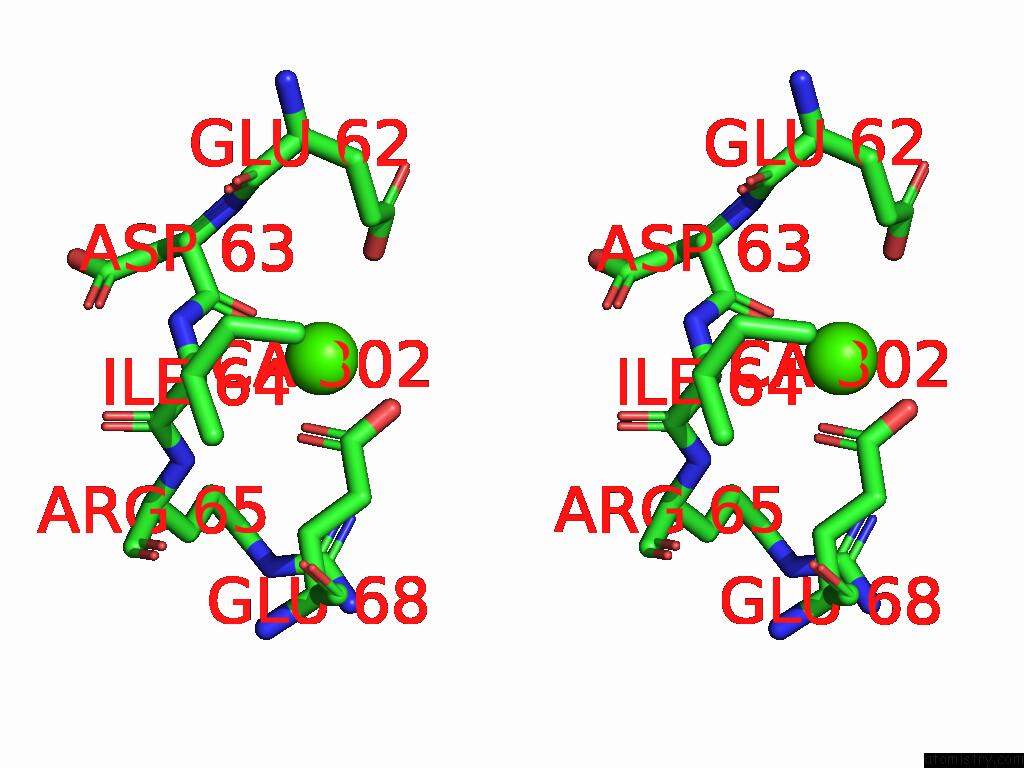
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Structure of Citrobacter Multi-Ubiquitin Protein, Local Refinement of One Full-Length Protomer within 5.0Å range:
|
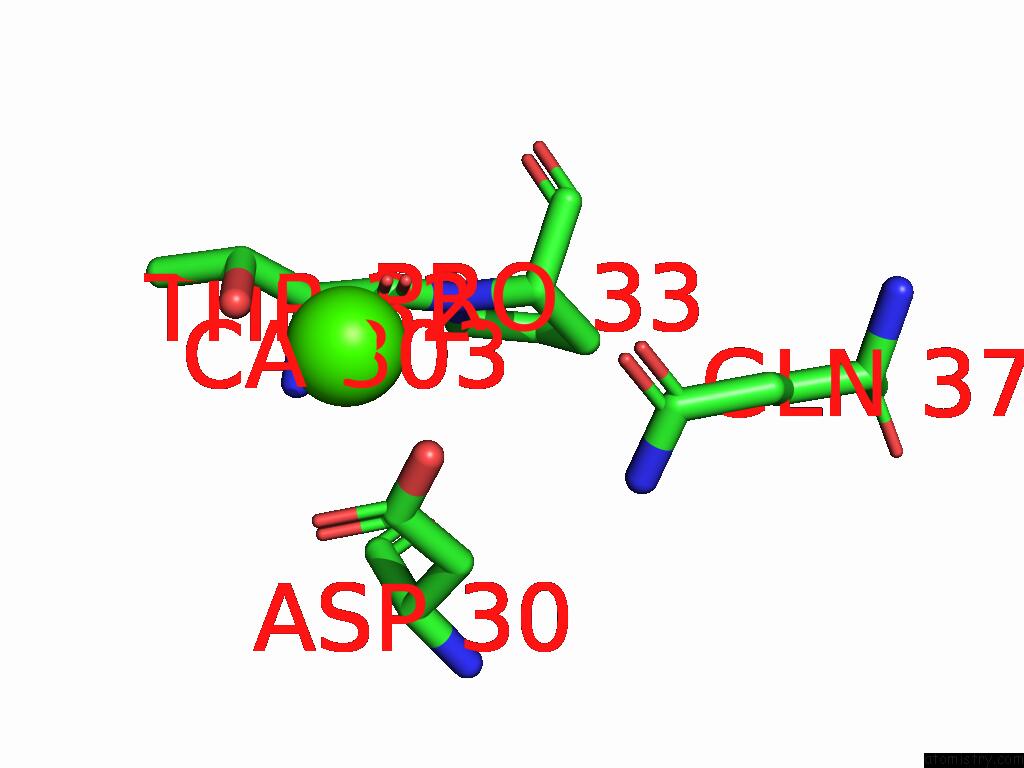
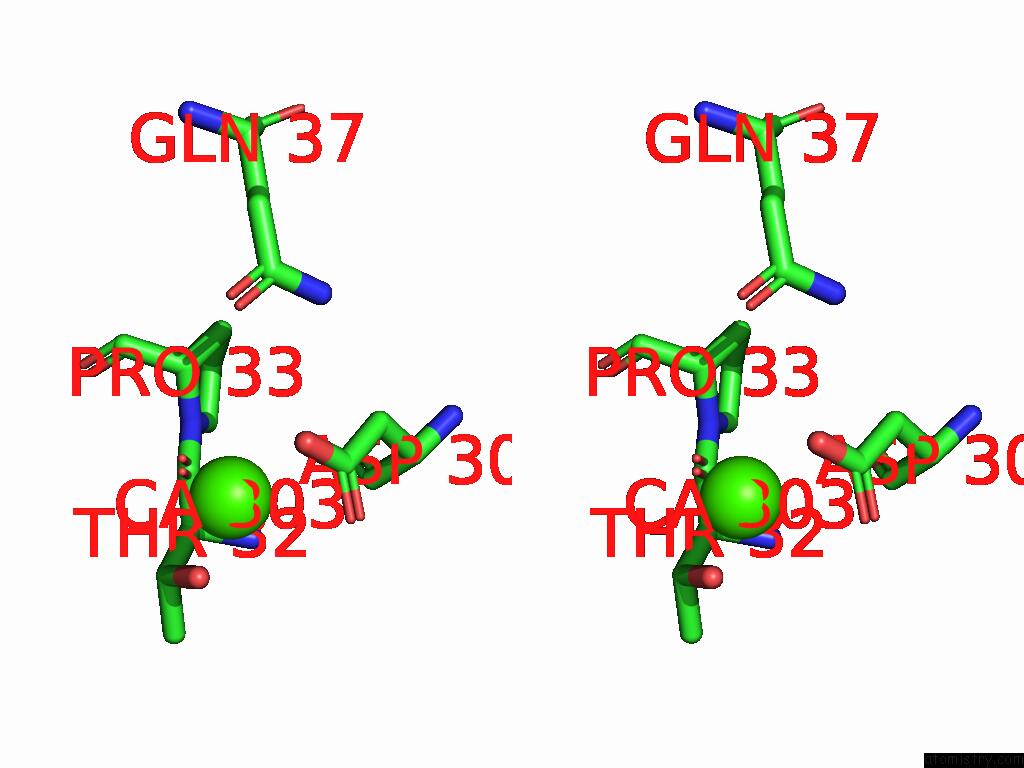
Calcium binding site 3 out of 5 in 9d5a
Go back to
Calcium binding site 3 out
of 5 in the Structure of Citrobacter Multi-Ubiquitin Protein, Local Refinement of One Full-Length Protomer
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 3 of Structure of Citrobacter Multi-Ubiquitin Protein, Local Refinement of One Full-Length Protomer within 5.0Å range:
|
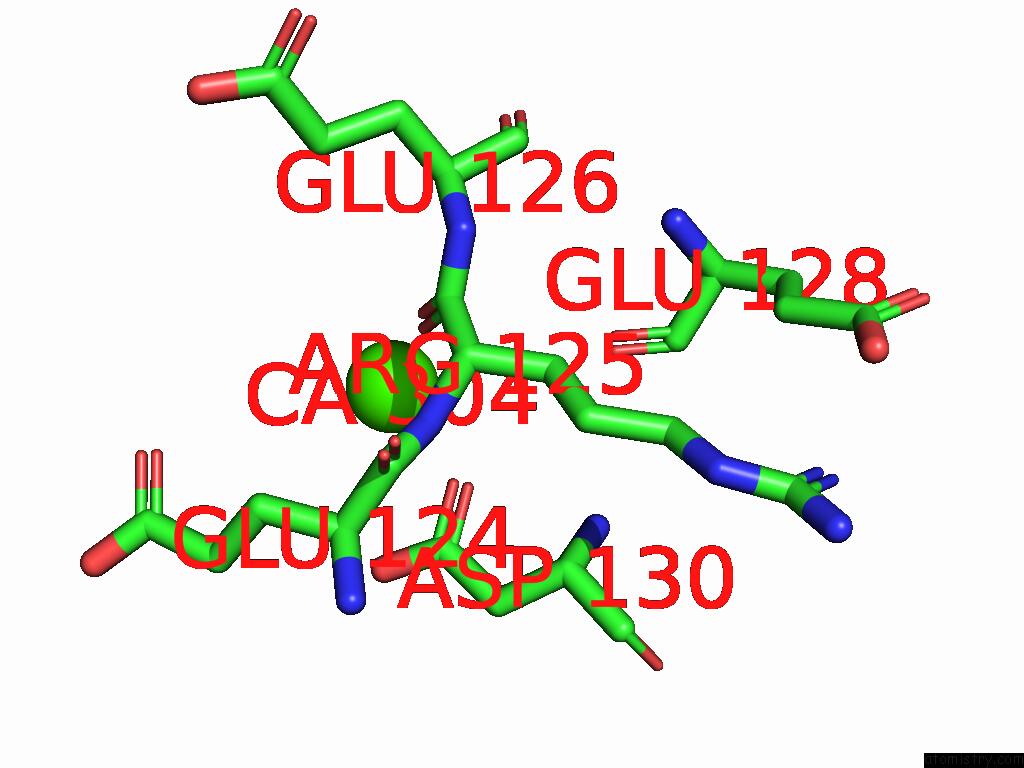
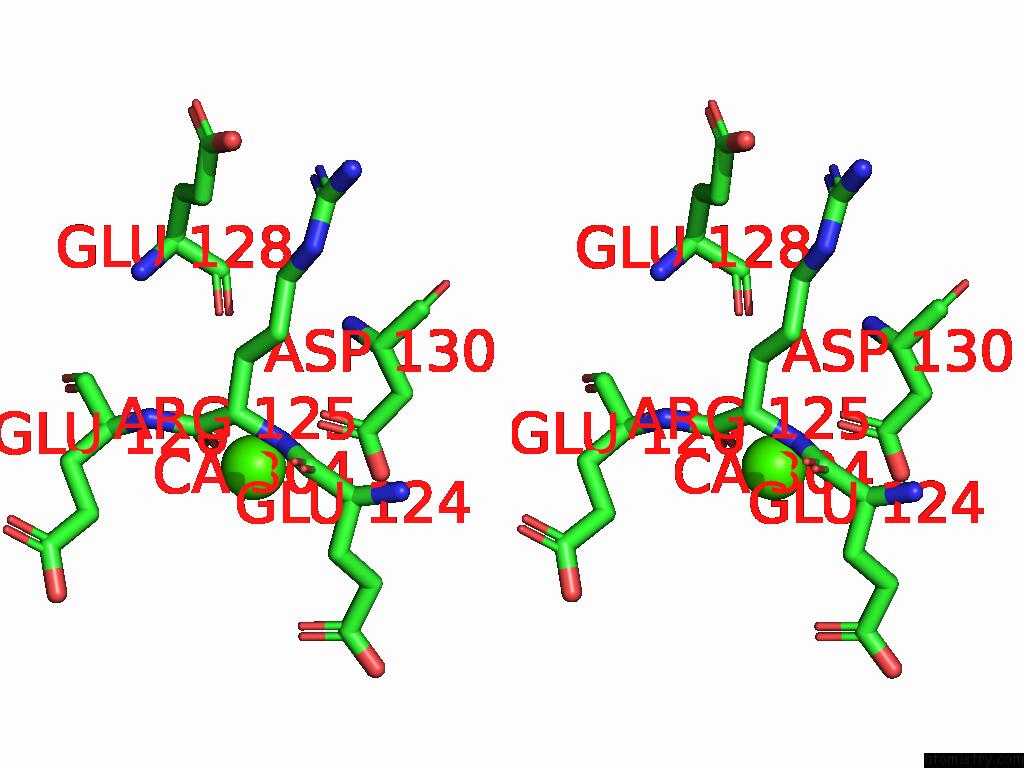
Calcium binding site 4 out of 5 in 9d5a
Go back to
Calcium binding site 4 out
of 5 in the Structure of Citrobacter Multi-Ubiquitin Protein, Local Refinement of One Full-Length Protomer
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 4 of Structure of Citrobacter Multi-Ubiquitin Protein, Local Refinement of One Full-Length Protomer within 5.0Å range:
|
Calcium binding site 5 out of 5 in 9d5a
Go back to
Calcium binding site 5 out
of 5 in the Structure of Citrobacter Multi-Ubiquitin Protein, Local Refinement of One Full-Length Protomer
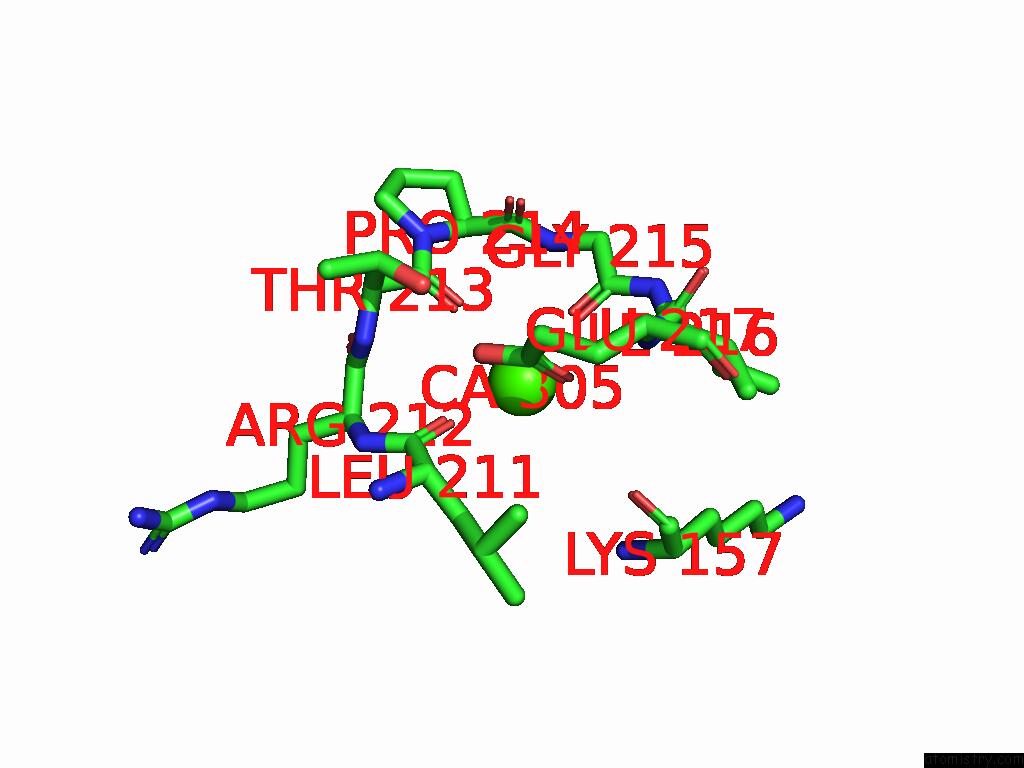
Mono view
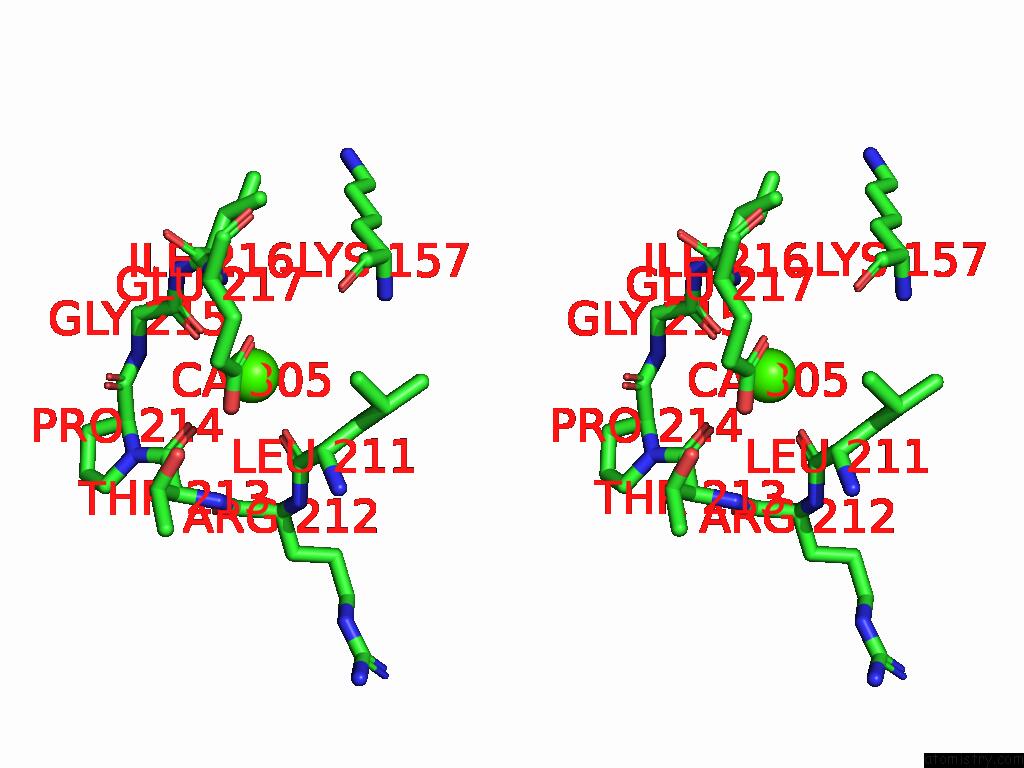
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 5 of Structure of Citrobacter Multi-Ubiquitin Protein, Local Refinement of One Full-Length Protomer within 5.0Å range:
|
Reference:
M.Gong,
Q.Ye,
Y.Gu,
L.R.Chambers,
M.Matyszewski,
K.D.Corbett.
Structural Diversity and Oligomerization of Bacterial Ubiquitin-Like Proteins To Be Published.
Page generated: Thu Jul 10 09:15:16 2025
Last articles
Fe in 8Q1UFe in 8Q2F
Fe in 8Q1W
Fe in 8Q1V
Fe in 8Q1P
Fe in 8Q0O
Fe in 8Q0Q
Fe in 8Q1B
Fe in 8Q0M
Fe in 8Q0F