Calcium »
PDB 9dqz-9eu5 »
9en2 »
Calcium in PDB 9en2: Crystal Structure of the Metalloproteinase Enhancer Pcpe-1 Complexed with Nanobodies Vhh-H4 and Vhh-I5
Protein crystallography data
The structure of Crystal Structure of the Metalloproteinase Enhancer Pcpe-1 Complexed with Nanobodies Vhh-H4 and Vhh-I5, PDB code: 9en2
was solved by
P.Lagoutte,
V.Gueguen-Chaignon,
J.-M.Bourhis,
S.Vadon-Le Goff,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 49.00 / 2.20 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 54.83, 95.945, 110.771, 90, 90, 90 |
| R / Rfree (%) | 20.8 / 24.2 |
Calcium Binding Sites:
The binding sites of Calcium atom in the Crystal Structure of the Metalloproteinase Enhancer Pcpe-1 Complexed with Nanobodies Vhh-H4 and Vhh-I5
(pdb code 9en2). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 2 binding sites of Calcium where determined in the Crystal Structure of the Metalloproteinase Enhancer Pcpe-1 Complexed with Nanobodies Vhh-H4 and Vhh-I5, PDB code: 9en2:
Jump to Calcium binding site number: 1; 2;
In total 2 binding sites of Calcium where determined in the Crystal Structure of the Metalloproteinase Enhancer Pcpe-1 Complexed with Nanobodies Vhh-H4 and Vhh-I5, PDB code: 9en2:
Jump to Calcium binding site number: 1; 2;
Calcium binding site 1 out of 2 in 9en2
Go back to
Calcium binding site 1 out
of 2 in the Crystal Structure of the Metalloproteinase Enhancer Pcpe-1 Complexed with Nanobodies Vhh-H4 and Vhh-I5
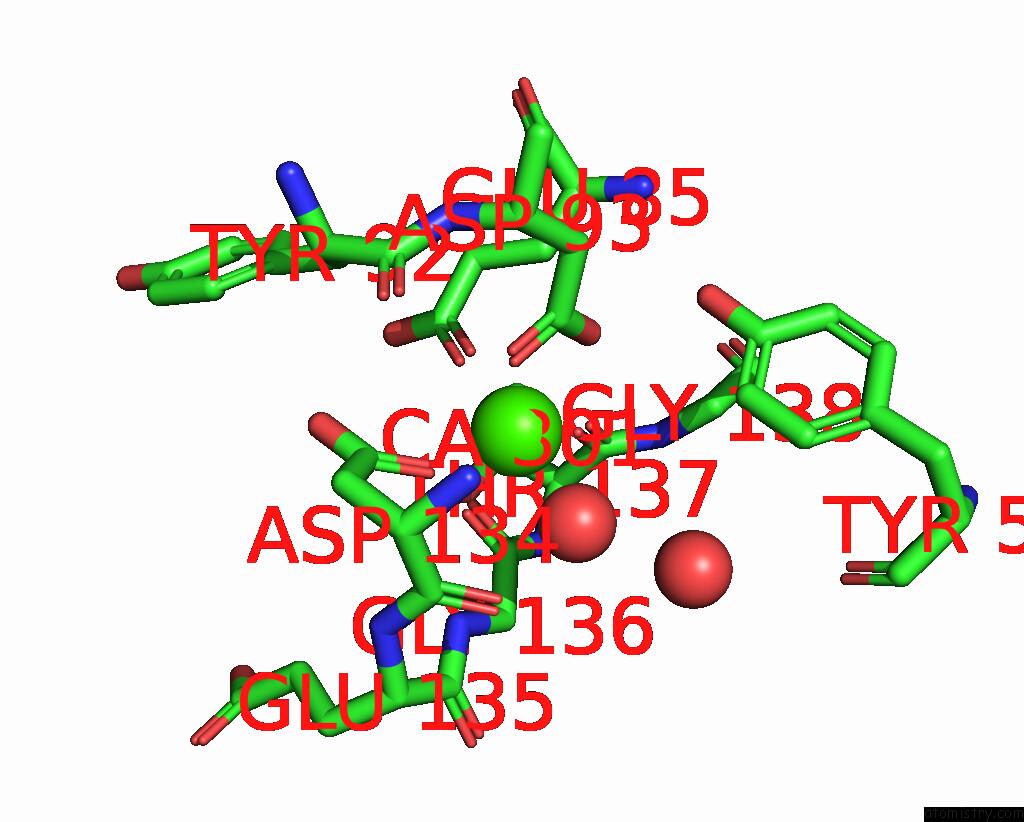
Mono view
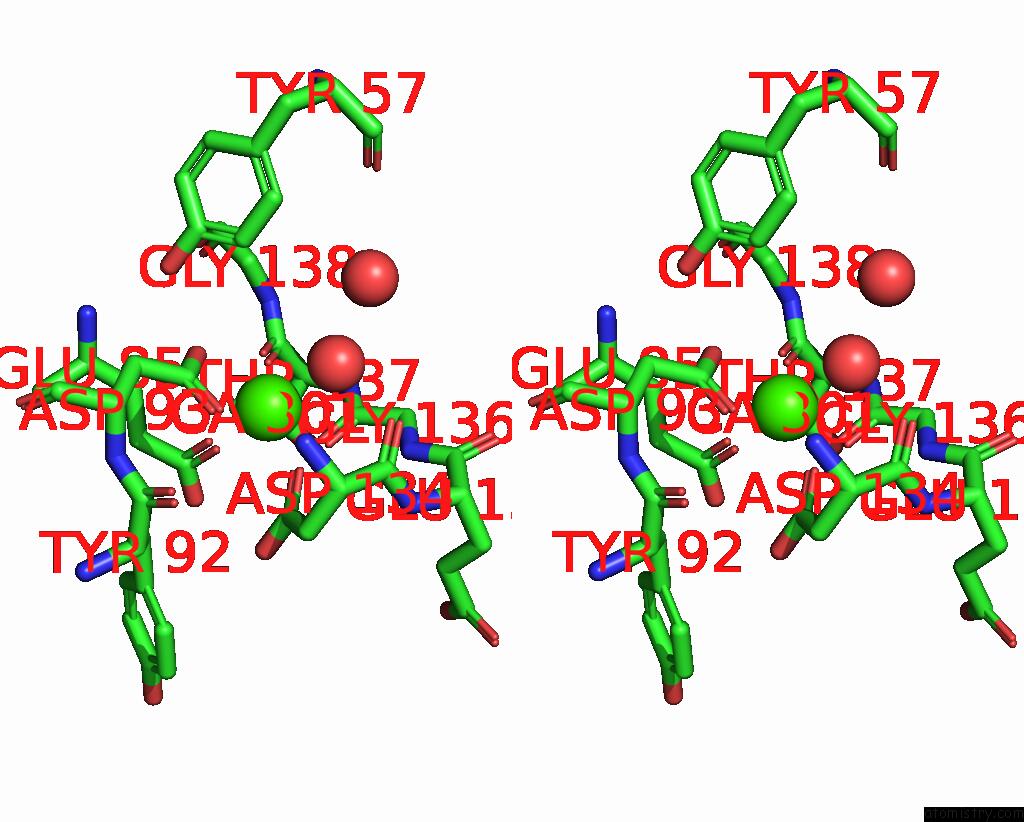
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Crystal Structure of the Metalloproteinase Enhancer Pcpe-1 Complexed with Nanobodies Vhh-H4 and Vhh-I5 within 5.0Å range:
|
Calcium binding site 2 out of 2 in 9en2
Go back to
Calcium binding site 2 out
of 2 in the Crystal Structure of the Metalloproteinase Enhancer Pcpe-1 Complexed with Nanobodies Vhh-H4 and Vhh-I5
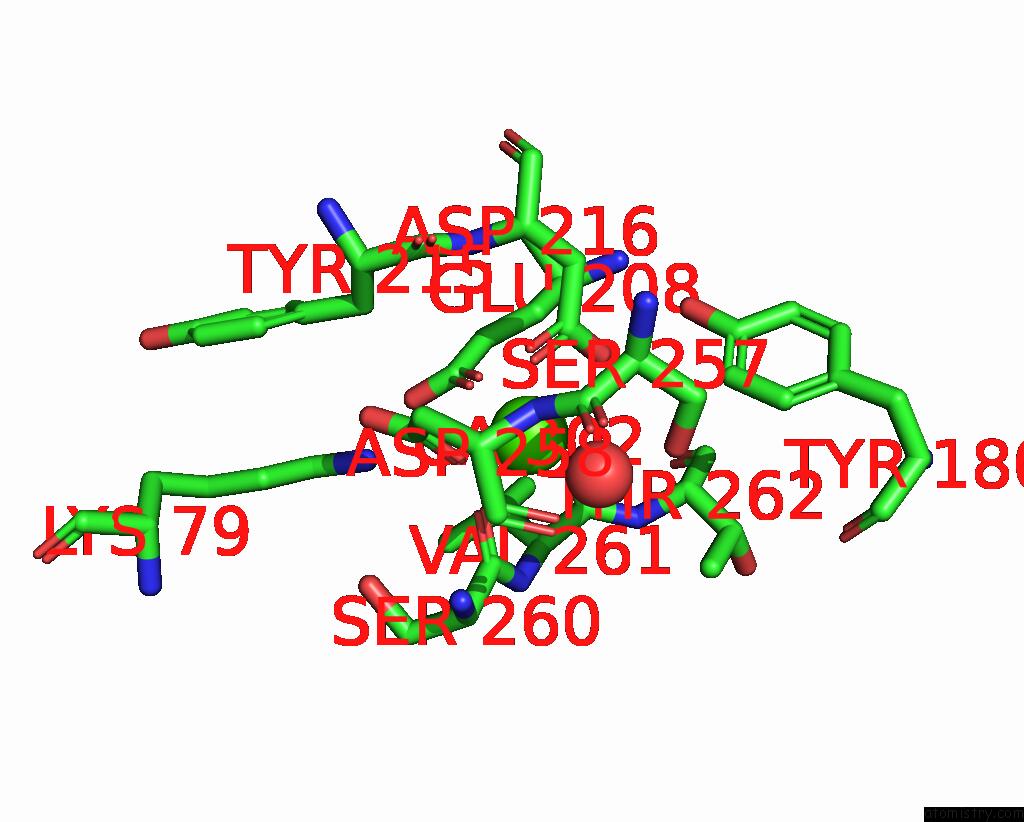
Mono view
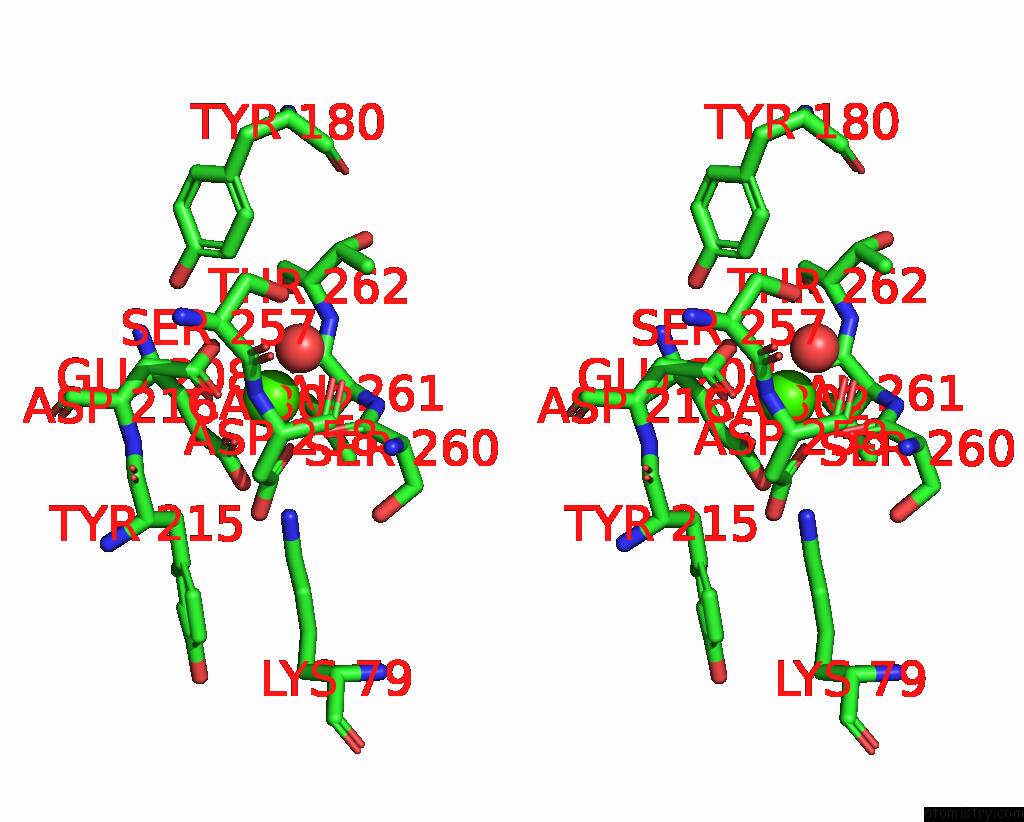
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Crystal Structure of the Metalloproteinase Enhancer Pcpe-1 Complexed with Nanobodies Vhh-H4 and Vhh-I5 within 5.0Å range:
|
Reference:
P.Lagoutte,
J.M.Bourhis,
N.Mariano,
V.Gueguen-Chaignon,
D.Vandroux,
C.Moali,
S.Vadon-Le Goff.
Mono- and Bi-Specific Nanobodies Targeting the Cub Domains of Pcpe-1 Reduce the Proteolytic Processing of Fibrillar Procollagens. J.Mol.Biol. 68667 2024.
ISSN: ESSN 1089-8638
PubMed: 38901640
DOI: 10.1016/J.JMB.2024.168667
Page generated: Thu Jul 10 09:32:50 2025
ISSN: ESSN 1089-8638
PubMed: 38901640
DOI: 10.1016/J.JMB.2024.168667
Last articles
Fe in 4L3ZFe in 4L4B
Fe in 4L3Y
Fe in 4L3X
Fe in 4L4A
Fe in 4L49
Fe in 4L40
Fe in 4L38
Fe in 4L3H
Fe in 4L3V