Calcium »
PDB 9fli-9gqx »
9ggc »
Calcium in PDB 9ggc: Structure of the G848S Mutant of Human Mitochondrial Dna Polymerase Gamma
Enzymatic activity of Structure of the G848S Mutant of Human Mitochondrial Dna Polymerase Gamma
All present enzymatic activity of Structure of the G848S Mutant of Human Mitochondrial Dna Polymerase Gamma:
2.7.7.7;
2.7.7.7;
Calcium Binding Sites:
The binding sites of Calcium atom in the Structure of the G848S Mutant of Human Mitochondrial Dna Polymerase Gamma
(pdb code 9ggc). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total only one binding site of Calcium was determined in the Structure of the G848S Mutant of Human Mitochondrial Dna Polymerase Gamma, PDB code: 9ggc:
In total only one binding site of Calcium was determined in the Structure of the G848S Mutant of Human Mitochondrial Dna Polymerase Gamma, PDB code: 9ggc:
Calcium binding site 1 out of 1 in 9ggc
Go back to
Calcium binding site 1 out
of 1 in the Structure of the G848S Mutant of Human Mitochondrial Dna Polymerase Gamma
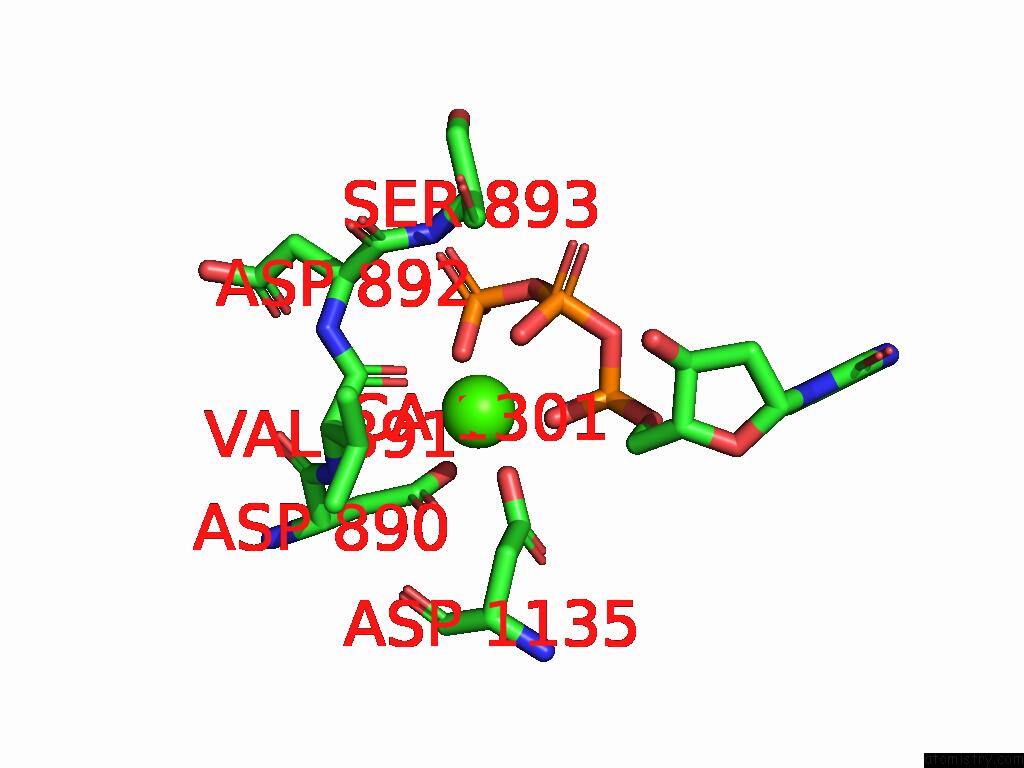
Mono view
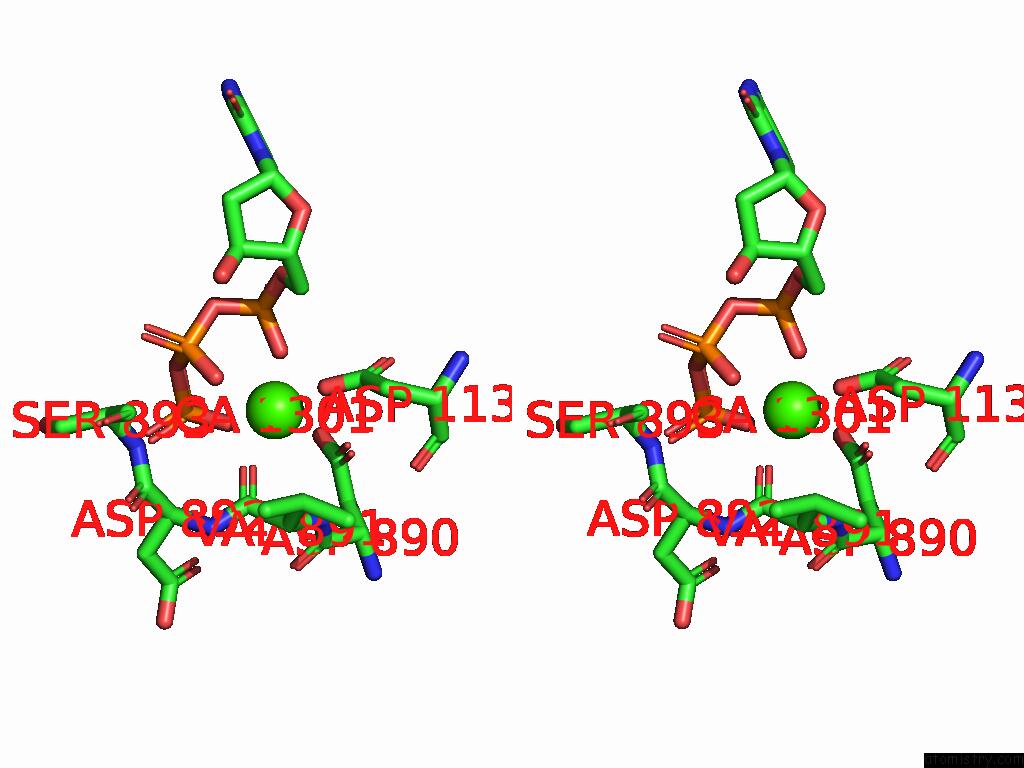
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Structure of the G848S Mutant of Human Mitochondrial Dna Polymerase Gamma within 5.0Å range:
|
Reference:
S.Valenzuela,
Z.Xuefeng,
M.Macao,
M.Stamgren,
C.Geukens,
P.S.Charifson,
G.Kern,
E.Hoberg,
L.Jenninger,
A.V.Gruszczyk,
S.Lee,
K.A.S.Johansson,
J.Miralles Fuste,
Y.Shi,
S.J.Kerns,
L.Arabanian,
G.Martinez Botella,
J.Green,
A.Griffin,
C.Pardo Hernandez,
T.A.Keating,
N.G.Larsson,
C.Phan,
V.Posse,
J.E.Jones,
X.Xie,
S.Giroux,
C.M.Gustafsson,
M.Falkenberg.
Small Molecules Restore Mutant Mitochondrial Dna Polymerase Activity To Be Published 2025.
DOI: 10.1038/S41586-025-08856-9
Page generated: Thu Jul 10 09:48:25 2025
DOI: 10.1038/S41586-025-08856-9
Last articles
Fe in 3OZWFe in 3OR2
Fe in 3OZV
Fe in 3OZU
Fe in 3OX4
Fe in 3OWO
Fe in 3OV0
Fe in 3OW5
Fe in 3OVU
Fe in 3OVR