Calcium »
PDB 9ibz-9kzc »
9ic3 »
Calcium in PDB 9ic3: Chimeric Mitochondrial Dna Polymerase Gamma Ternary Complex (Hamb) in Mouse-Like Error-Editing Conformer (Composite)
Enzymatic activity of Chimeric Mitochondrial Dna Polymerase Gamma Ternary Complex (Hamb) in Mouse-Like Error-Editing Conformer (Composite)
All present enzymatic activity of Chimeric Mitochondrial Dna Polymerase Gamma Ternary Complex (Hamb) in Mouse-Like Error-Editing Conformer (Composite):
2.7.7.7;
2.7.7.7;
Calcium Binding Sites:
The binding sites of Calcium atom in the Chimeric Mitochondrial Dna Polymerase Gamma Ternary Complex (Hamb) in Mouse-Like Error-Editing Conformer (Composite)
(pdb code 9ic3). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 3 binding sites of Calcium where determined in the Chimeric Mitochondrial Dna Polymerase Gamma Ternary Complex (Hamb) in Mouse-Like Error-Editing Conformer (Composite), PDB code: 9ic3:
Jump to Calcium binding site number: 1; 2; 3;
In total 3 binding sites of Calcium where determined in the Chimeric Mitochondrial Dna Polymerase Gamma Ternary Complex (Hamb) in Mouse-Like Error-Editing Conformer (Composite), PDB code: 9ic3:
Jump to Calcium binding site number: 1; 2; 3;
Calcium binding site 1 out of 3 in 9ic3
Go back to
Calcium binding site 1 out
of 3 in the Chimeric Mitochondrial Dna Polymerase Gamma Ternary Complex (Hamb) in Mouse-Like Error-Editing Conformer (Composite)
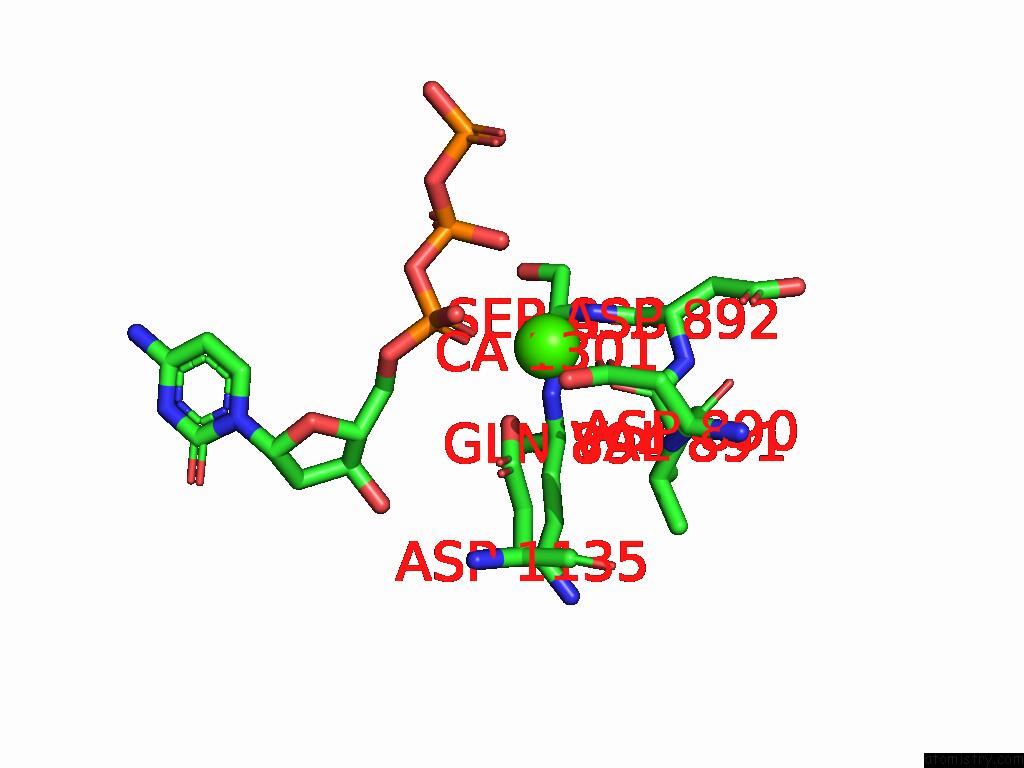
Mono view
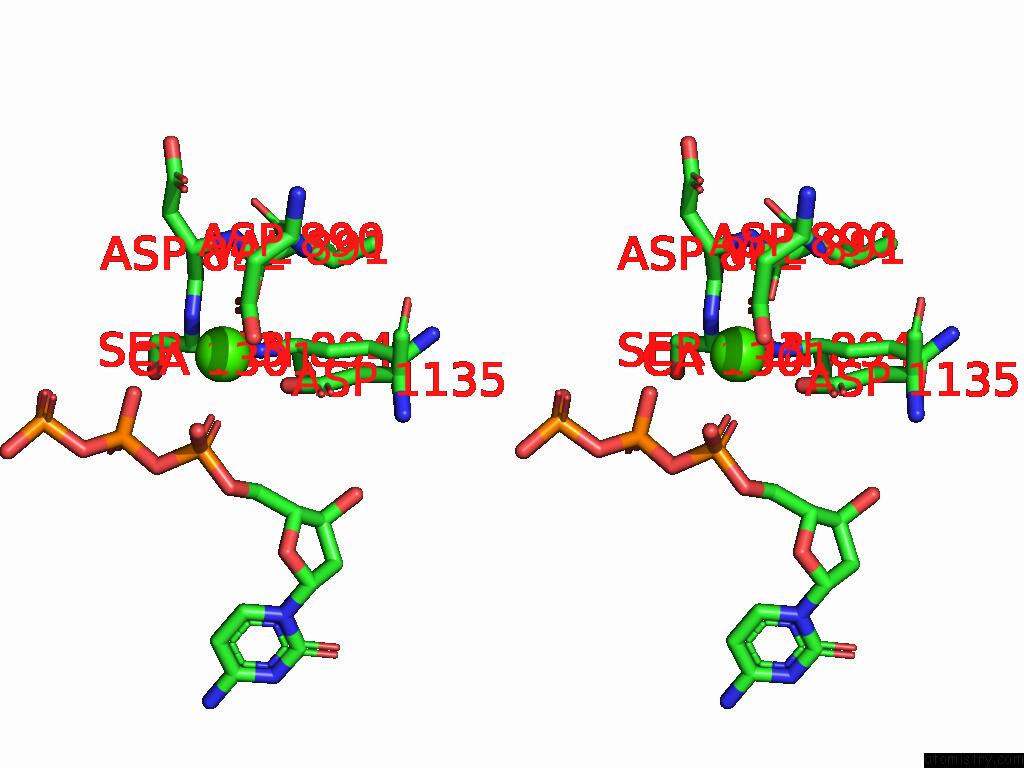
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Chimeric Mitochondrial Dna Polymerase Gamma Ternary Complex (Hamb) in Mouse-Like Error-Editing Conformer (Composite) within 5.0Å range:
|
Calcium binding site 2 out of 3 in 9ic3
Go back to
Calcium binding site 2 out
of 3 in the Chimeric Mitochondrial Dna Polymerase Gamma Ternary Complex (Hamb) in Mouse-Like Error-Editing Conformer (Composite)
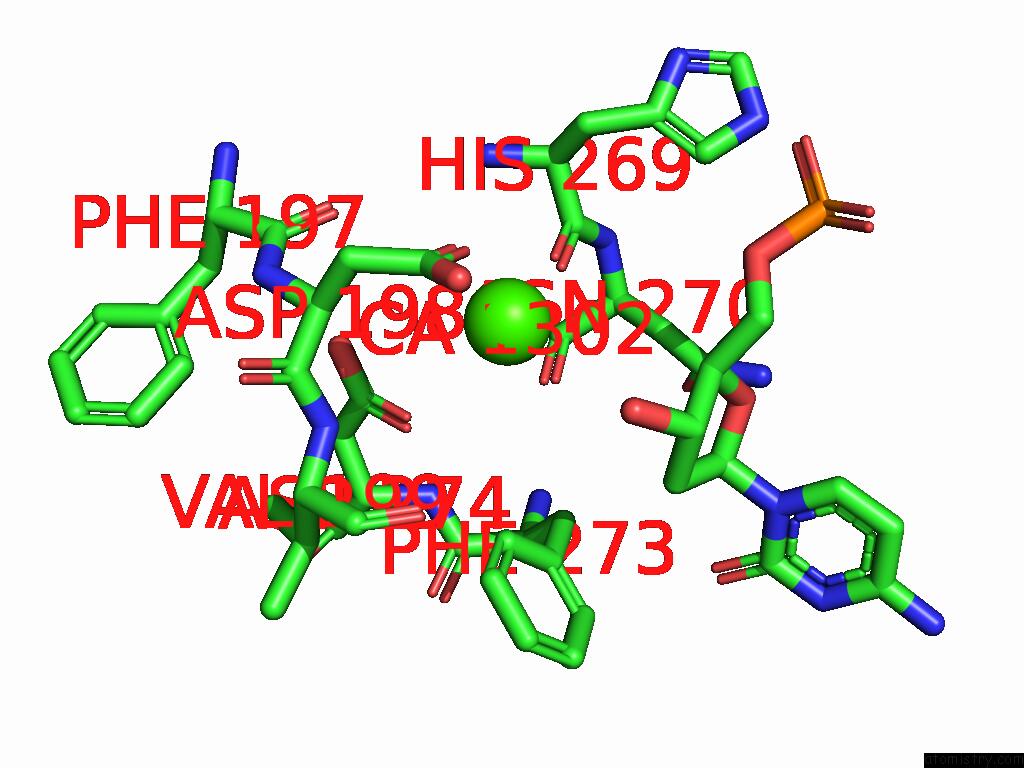
Mono view
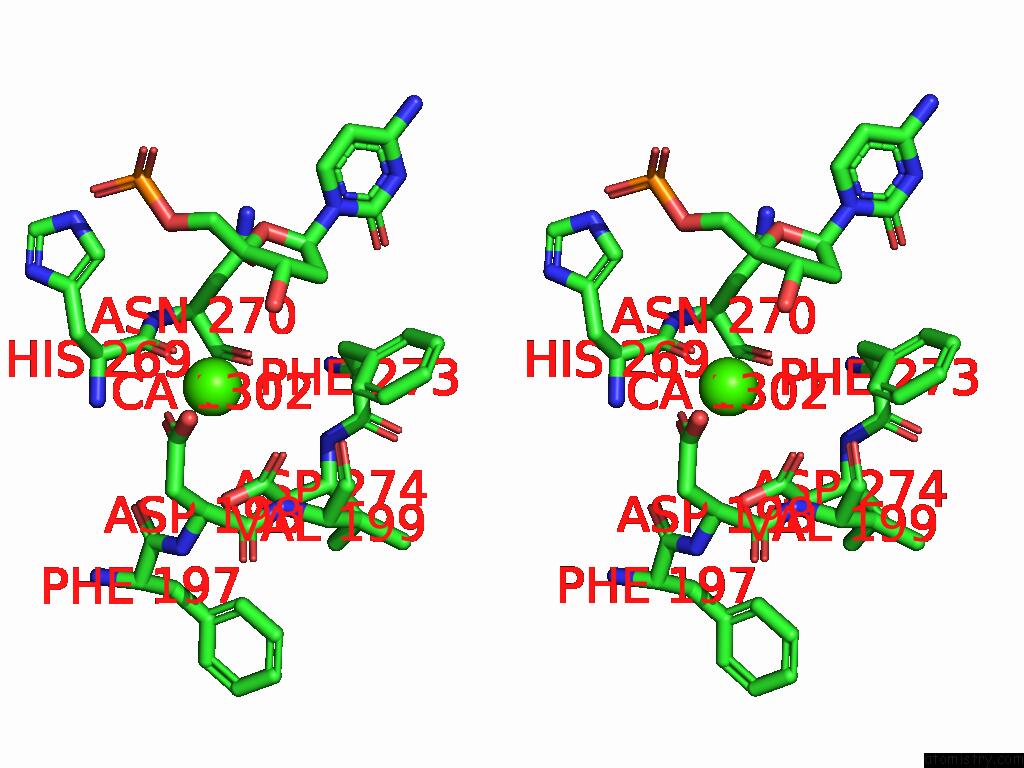
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Chimeric Mitochondrial Dna Polymerase Gamma Ternary Complex (Hamb) in Mouse-Like Error-Editing Conformer (Composite) within 5.0Å range:
|
Calcium binding site 3 out of 3 in 9ic3
Go back to
Calcium binding site 3 out
of 3 in the Chimeric Mitochondrial Dna Polymerase Gamma Ternary Complex (Hamb) in Mouse-Like Error-Editing Conformer (Composite)
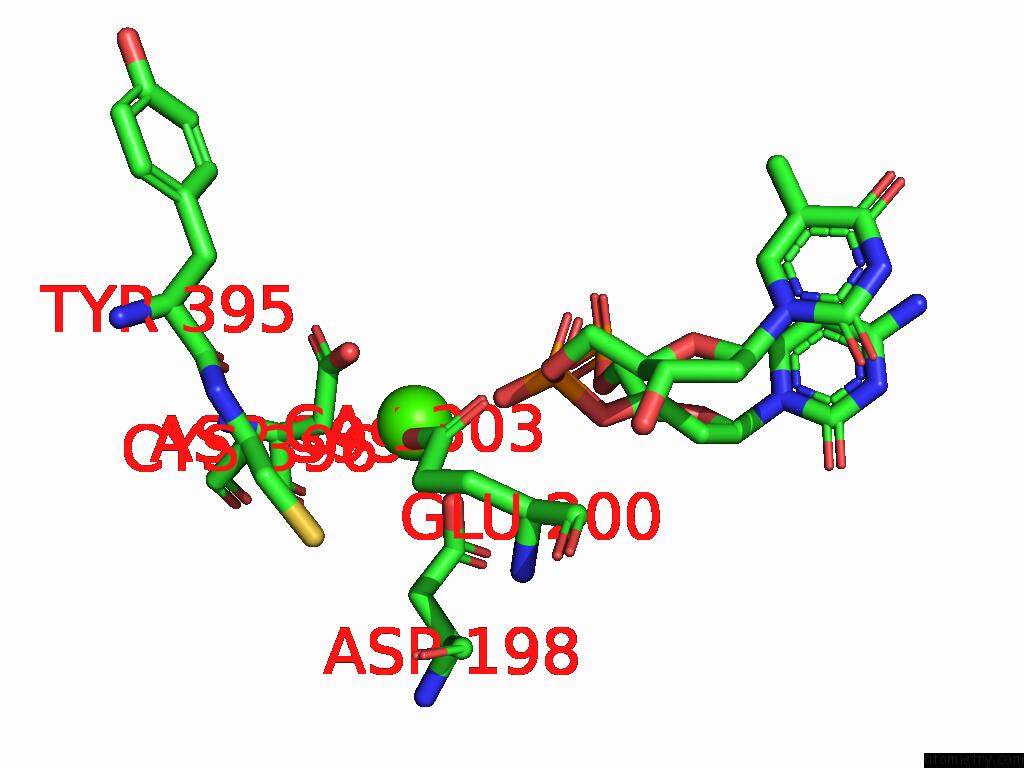
Mono view
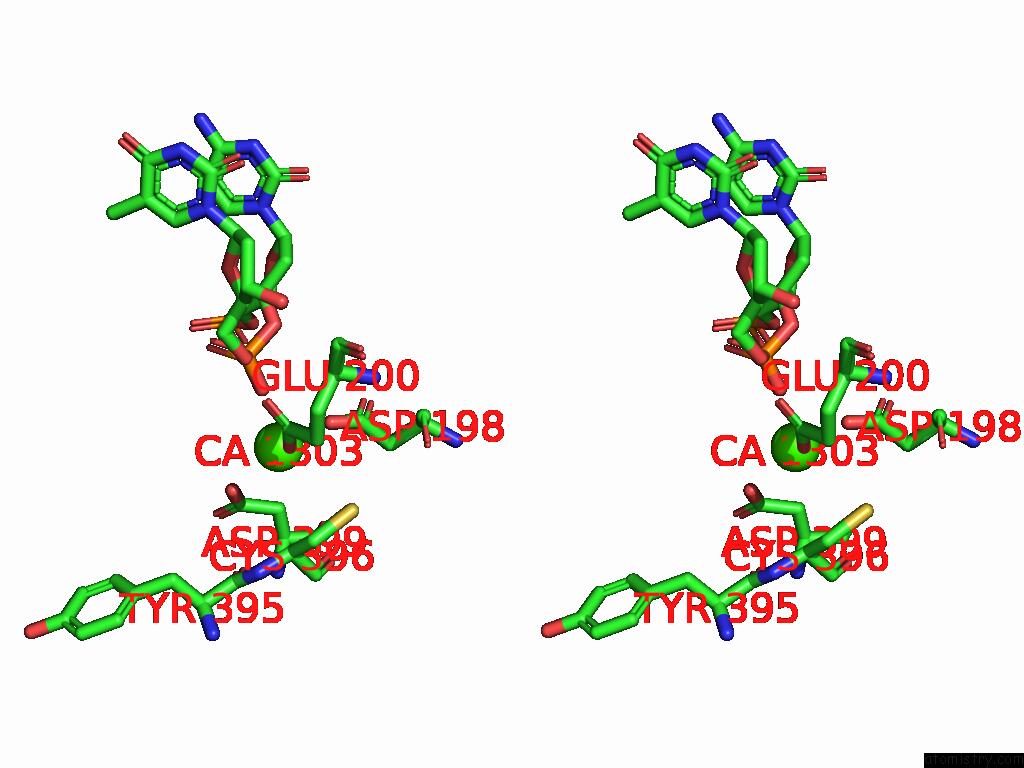
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 3 of Chimeric Mitochondrial Dna Polymerase Gamma Ternary Complex (Hamb) in Mouse-Like Error-Editing Conformer (Composite) within 5.0Å range:
|
Reference:
S.Corra,
A.Zuppardo,
S.Valenzuela,
L.Jenninger,
R.Cerutti,
S.Sillamaa,
E.Hoberg,
K.A.S.Johansson,
U.Rovsnik,
S.Volta,
P.Silva-Pinheiro,
H.Davis,
A.Trifunovic,
M.Minczuk,
C.M.Gustafsson,
A.Suomalainen,
M.Zeviani,
B.Macao,
X.Zhu,
M.Falkenberg,
C.Viscomi.
Modelling Polg Mutations in Mice Unravels A Critical Role of Pol Gamma Beta in Regulating Phenotypic Severity. Nat Commun V. 16 4782 2025.
ISSN: ESSN 2041-1723
PubMed: 40404629
DOI: 10.1038/S41467-025-60059-Y
Page generated: Thu Jul 10 09:54:54 2025
ISSN: ESSN 2041-1723
PubMed: 40404629
DOI: 10.1038/S41467-025-60059-Y
Last articles
Fe in 2YXOFe in 2YRS
Fe in 2YXC
Fe in 2YNM
Fe in 2YVJ
Fe in 2YP1
Fe in 2YU2
Fe in 2YU1
Fe in 2YQB
Fe in 2YOO