Calcium »
PDB 2fod-2g81 »
2g4j »
Calcium in PDB 2g4j: Anomalous Substructure of Glucose Isomerase
Enzymatic activity of Anomalous Substructure of Glucose Isomerase
All present enzymatic activity of Anomalous Substructure of Glucose Isomerase:
5.3.1.5;
5.3.1.5;
Protein crystallography data
The structure of Anomalous Substructure of Glucose Isomerase, PDB code: 2g4j
was solved by
C.Mueller-Dieckmann,
M.S.Weiss,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 30.00 / 1.85 |
| Space group | I 2 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 93.020, 97.590, 102.770, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 15.2 / 17.7 |
Other elements in 2g4j:
The structure of Anomalous Substructure of Glucose Isomerase also contains other interesting chemical elements:
| Magnesium | (Mg) | 1 atom |
| Chlorine | (Cl) | 1 atom |
Calcium Binding Sites:
The binding sites of Calcium atom in the Anomalous Substructure of Glucose Isomerase
(pdb code 2g4j). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total only one binding site of Calcium was determined in the Anomalous Substructure of Glucose Isomerase, PDB code: 2g4j:
In total only one binding site of Calcium was determined in the Anomalous Substructure of Glucose Isomerase, PDB code: 2g4j:
Calcium binding site 1 out of 1 in 2g4j
Go back to
Calcium binding site 1 out
of 1 in the Anomalous Substructure of Glucose Isomerase
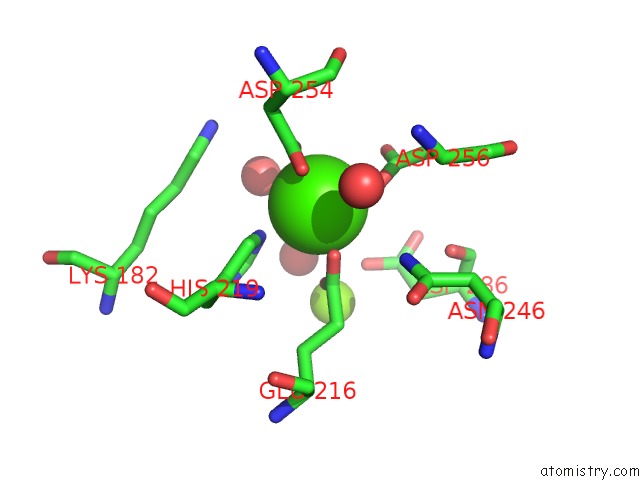
Mono view
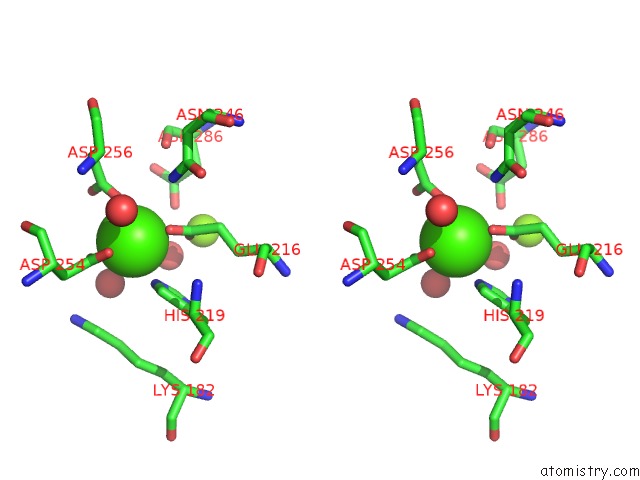
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Anomalous Substructure of Glucose Isomerase within 5.0Å range:
|
Reference:
C.Mueller-Dieckmann,
S.Panjikar,
A.Schmidt,
S.Mueller,
J.Kuper,
A.Geerlof,
M.Wilmanns,
R.K.Singh,
P.A.Tucker,
M.S.Weiss.
On the Routine Use of Soft X-Rays in Macromolecular Crystallography. Part IV. Efficient Determination of Anomalous Substructures in Biomacromolecules Using Longer X-Ray Wavelengths. Acta Crystallogr.,Sect.D V. 63 366 2007.
ISSN: ISSN 0907-4449
PubMed: 17327674
DOI: 10.1107/S0907444906055624
Page generated: Tue Jul 8 05:40:54 2025
ISSN: ISSN 0907-4449
PubMed: 17327674
DOI: 10.1107/S0907444906055624
Last articles
Ca in 7EV1Ca in 7F44
Ca in 7F06
Ca in 7EZZ
Ca in 7EY3
Ca in 7EXC
Ca in 7EXK
Ca in 7EXO
Ca in 7ETI
Ca in 7ETH