Calcium »
PDB 2kug-2m0j »
2l7l »
Calcium in PDB 2l7l: Solution Structure of CA2+/Calmodulin Complexed with A Peptide Representing the Calmodulin-Binding Domain of Calmodulin Kinase I
Enzymatic activity of Solution Structure of CA2+/Calmodulin Complexed with A Peptide Representing the Calmodulin-Binding Domain of Calmodulin Kinase I
All present enzymatic activity of Solution Structure of CA2+/Calmodulin Complexed with A Peptide Representing the Calmodulin-Binding Domain of Calmodulin Kinase I:
2.7.11.17;
2.7.11.17;
Calcium Binding Sites:
The binding sites of Calcium atom in the Solution Structure of CA2+/Calmodulin Complexed with A Peptide Representing the Calmodulin-Binding Domain of Calmodulin Kinase I
(pdb code 2l7l). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 4 binding sites of Calcium where determined in the Solution Structure of CA2+/Calmodulin Complexed with A Peptide Representing the Calmodulin-Binding Domain of Calmodulin Kinase I, PDB code: 2l7l:
Jump to Calcium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Calcium where determined in the Solution Structure of CA2+/Calmodulin Complexed with A Peptide Representing the Calmodulin-Binding Domain of Calmodulin Kinase I, PDB code: 2l7l:
Jump to Calcium binding site number: 1; 2; 3; 4;
Calcium binding site 1 out of 4 in 2l7l
Go back to
Calcium binding site 1 out
of 4 in the Solution Structure of CA2+/Calmodulin Complexed with A Peptide Representing the Calmodulin-Binding Domain of Calmodulin Kinase I
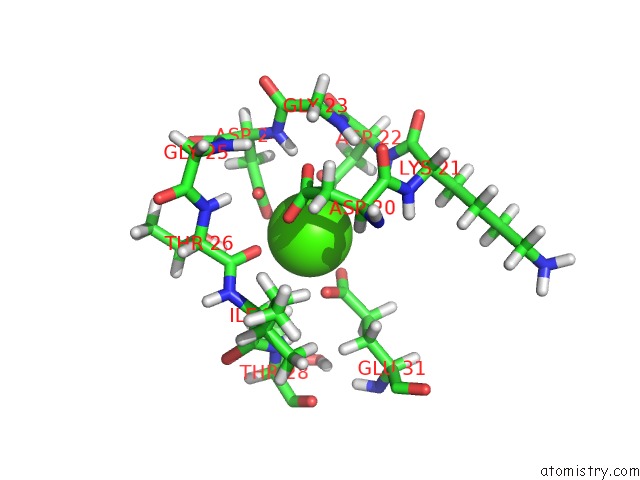
Mono view
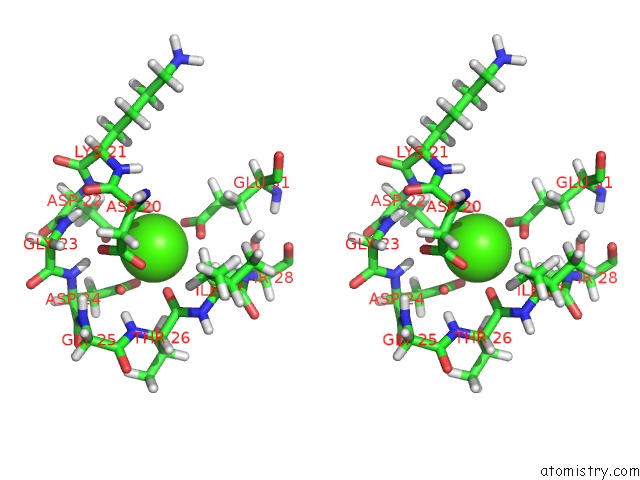
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Solution Structure of CA2+/Calmodulin Complexed with A Peptide Representing the Calmodulin-Binding Domain of Calmodulin Kinase I within 5.0Å range:
|
Calcium binding site 2 out of 4 in 2l7l
Go back to
Calcium binding site 2 out
of 4 in the Solution Structure of CA2+/Calmodulin Complexed with A Peptide Representing the Calmodulin-Binding Domain of Calmodulin Kinase I
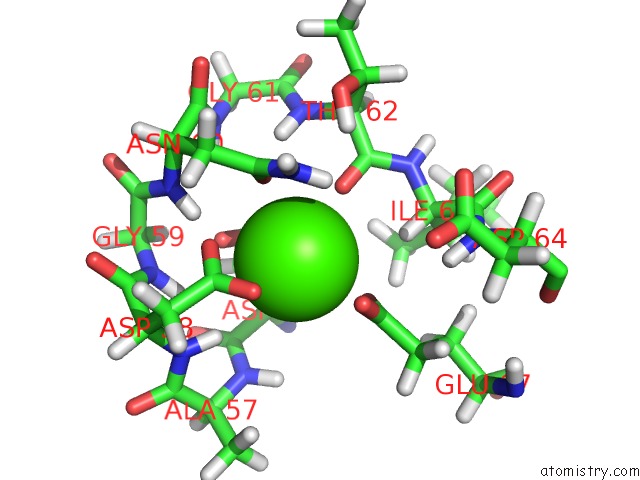
Mono view
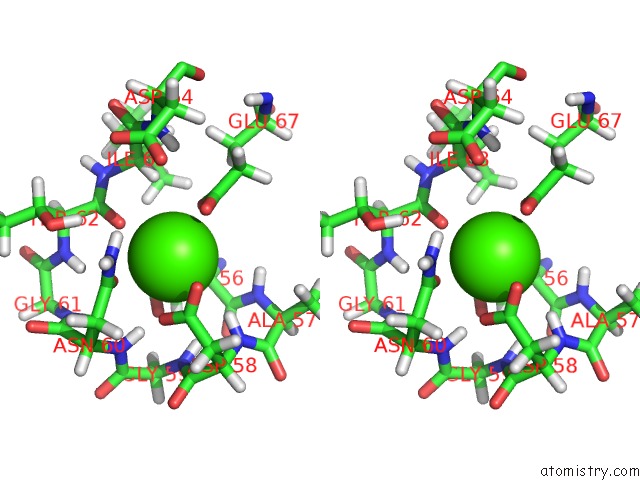
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Solution Structure of CA2+/Calmodulin Complexed with A Peptide Representing the Calmodulin-Binding Domain of Calmodulin Kinase I within 5.0Å range:
|
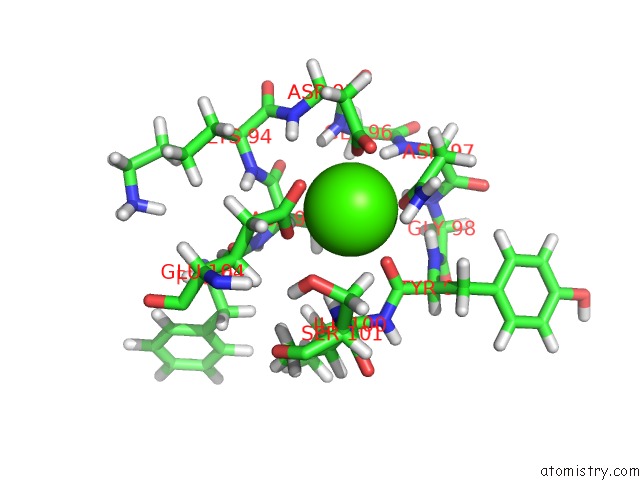
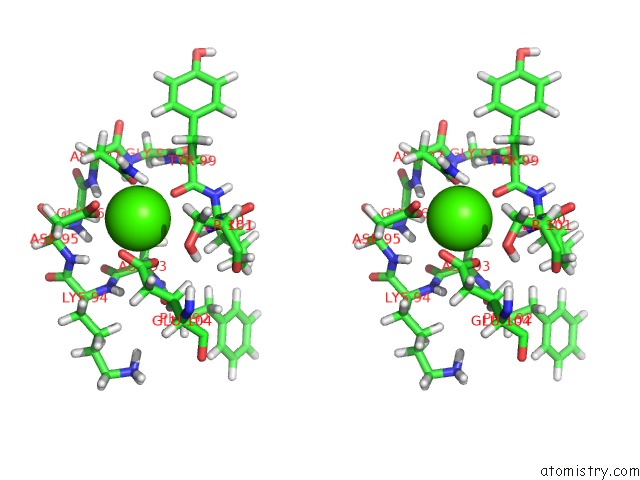
Calcium binding site 3 out of 4 in 2l7l
Go back to
Calcium binding site 3 out
of 4 in the Solution Structure of CA2+/Calmodulin Complexed with A Peptide Representing the Calmodulin-Binding Domain of Calmodulin Kinase I
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 3 of Solution Structure of CA2+/Calmodulin Complexed with A Peptide Representing the Calmodulin-Binding Domain of Calmodulin Kinase I within 5.0Å range:
|
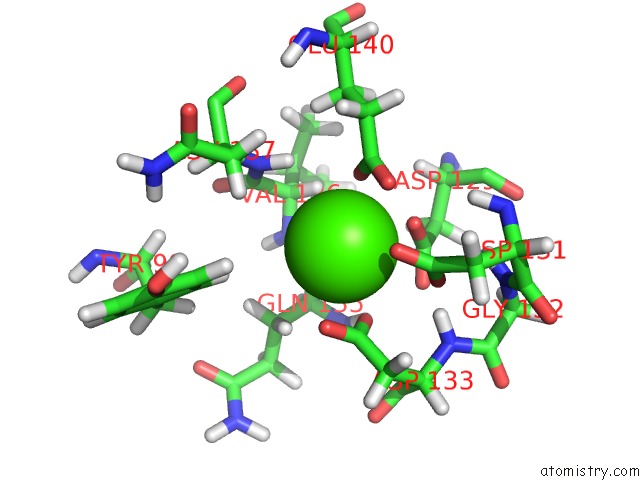
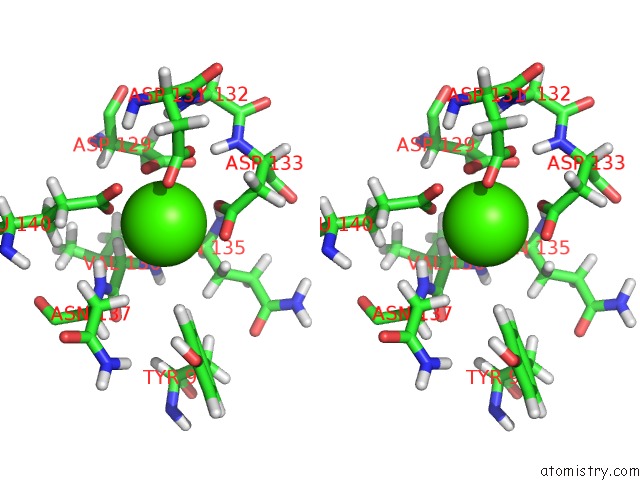
Calcium binding site 4 out of 4 in 2l7l
Go back to
Calcium binding site 4 out
of 4 in the Solution Structure of CA2+/Calmodulin Complexed with A Peptide Representing the Calmodulin-Binding Domain of Calmodulin Kinase I
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 4 of Solution Structure of CA2+/Calmodulin Complexed with A Peptide Representing the Calmodulin-Binding Domain of Calmodulin Kinase I within 5.0Å range:
|
Reference:
J.L.Gifford,
H.Ishida,
H.J.Vogel.
Fast Methionine-Based Solution Structure Determination of Calcium-Calmodulin Complexes. J.Biomol.uc(Nmr) V. 50 71 2011.
ISSN: ISSN 0925-2738
PubMed: 21360154
DOI: 10.1007/S10858-011-9495-3
Page generated: Tue Jul 8 06:55:03 2025
ISSN: ISSN 0925-2738
PubMed: 21360154
DOI: 10.1007/S10858-011-9495-3
Last articles
Ca in 3B8YCa in 3B7Y
Ca in 3B4N
Ca in 3B55
Ca in 3B40
Ca in 3B1U
Ca in 3B3Q
Ca in 3B3D
Ca in 3B32
Ca in 3B1T