Calcium »
PDB 2kug-2m0j »
2llq »
Calcium in PDB 2llq: Solution uc(Nmr)-Derived Structure of Calmodulin C-Lobe Bound with Er Alpha Peptide
Calcium Binding Sites:
The binding sites of Calcium atom in the Solution uc(Nmr)-Derived Structure of Calmodulin C-Lobe Bound with Er Alpha Peptide
(pdb code 2llq). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 2 binding sites of Calcium where determined in the Solution uc(Nmr)-Derived Structure of Calmodulin C-Lobe Bound with Er Alpha Peptide, PDB code: 2llq:
Jump to Calcium binding site number: 1; 2;
In total 2 binding sites of Calcium where determined in the Solution uc(Nmr)-Derived Structure of Calmodulin C-Lobe Bound with Er Alpha Peptide, PDB code: 2llq:
Jump to Calcium binding site number: 1; 2;
Calcium binding site 1 out of 2 in 2llq
Go back to
Calcium binding site 1 out
of 2 in the Solution uc(Nmr)-Derived Structure of Calmodulin C-Lobe Bound with Er Alpha Peptide
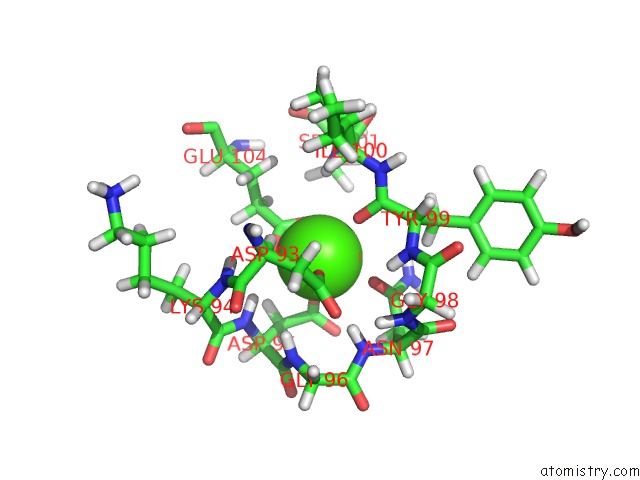
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Solution uc(Nmr)-Derived Structure of Calmodulin C-Lobe Bound with Er Alpha Peptide within 5.0Å range:
|
Calcium binding site 2 out of 2 in 2llq
Go back to
Calcium binding site 2 out
of 2 in the Solution uc(Nmr)-Derived Structure of Calmodulin C-Lobe Bound with Er Alpha Peptide
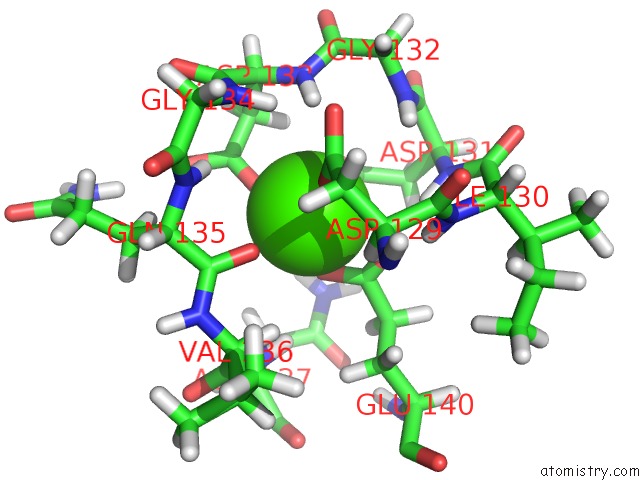
Mono view
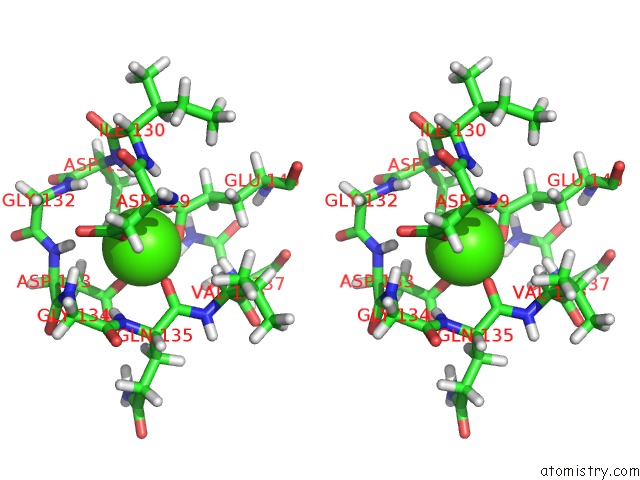
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Solution uc(Nmr)-Derived Structure of Calmodulin C-Lobe Bound with Er Alpha Peptide within 5.0Å range:
|
Reference:
Y.Zhang,
Z.Li,
D.B.Sacks,
J.B.Ames.
Structural Basis For CA2+-Induced Activation and Dimerization of Estrogen Receptor Alpha By Calmodulin. J.Biol.Chem. V. 287 9336 2012.
ISSN: ISSN 0021-9258
PubMed: 22275375
DOI: 10.1074/JBC.M111.334797
Page generated: Tue Jul 8 06:58:06 2025
ISSN: ISSN 0021-9258
PubMed: 22275375
DOI: 10.1074/JBC.M111.334797
Last articles
Ca in 3G5CCa in 3G6J
Ca in 3G5I
Ca in 3G5R
Ca in 3G4G
Ca in 3G4E
Ca in 3G20
Ca in 3FZ3
Ca in 3G27
Ca in 3FZ0