Calcium »
PDB 3aau-3ati »
3aqj »
Calcium in PDB 3aqj: Crystal Structure of A C-Terminal Domain of the Bacteriophage P2 Tail Spike Protein, Gpv
Protein crystallography data
The structure of Crystal Structure of A C-Terminal Domain of the Bacteriophage P2 Tail Spike Protein, Gpv, PDB code: 3aqj
was solved by
S.Takeda,
E.Yamashita,
A.Nakagawa,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 46.42 / 1.27 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 66.912, 64.463, 67.186, 90.00, 91.32, 90.00 |
| R / Rfree (%) | 16 / 19 |
Other elements in 3aqj:
The structure of Crystal Structure of A C-Terminal Domain of the Bacteriophage P2 Tail Spike Protein, Gpv also contains other interesting chemical elements:
| Chlorine | (Cl) | 2 atoms |
| Iron | (Fe) | 2 atoms |
Calcium Binding Sites:
The binding sites of Calcium atom in the Crystal Structure of A C-Terminal Domain of the Bacteriophage P2 Tail Spike Protein, Gpv
(pdb code 3aqj). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 2 binding sites of Calcium where determined in the Crystal Structure of A C-Terminal Domain of the Bacteriophage P2 Tail Spike Protein, Gpv, PDB code: 3aqj:
Jump to Calcium binding site number: 1; 2;
In total 2 binding sites of Calcium where determined in the Crystal Structure of A C-Terminal Domain of the Bacteriophage P2 Tail Spike Protein, Gpv, PDB code: 3aqj:
Jump to Calcium binding site number: 1; 2;
Calcium binding site 1 out of 2 in 3aqj
Go back to
Calcium binding site 1 out
of 2 in the Crystal Structure of A C-Terminal Domain of the Bacteriophage P2 Tail Spike Protein, Gpv
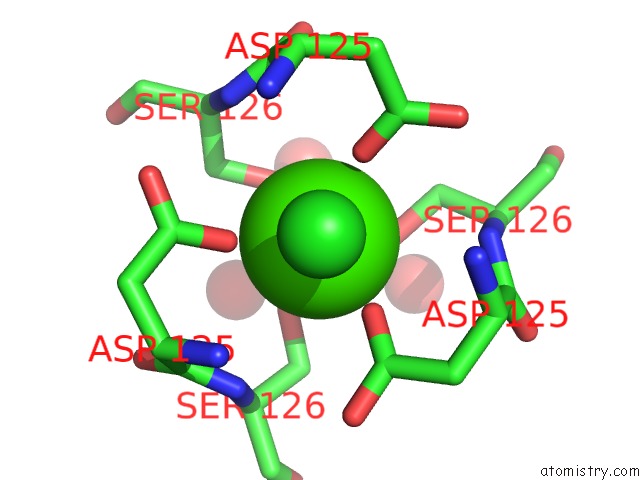
Mono view
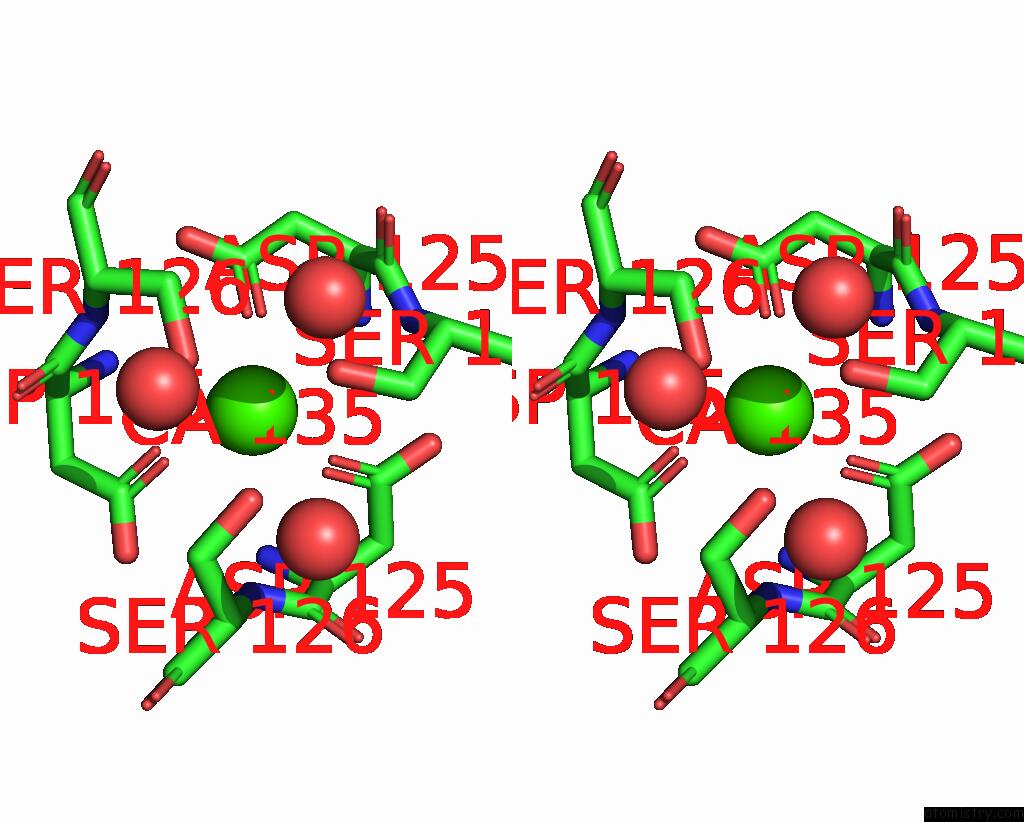
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Crystal Structure of A C-Terminal Domain of the Bacteriophage P2 Tail Spike Protein, Gpv within 5.0Å range:
|
Calcium binding site 2 out of 2 in 3aqj
Go back to
Calcium binding site 2 out
of 2 in the Crystal Structure of A C-Terminal Domain of the Bacteriophage P2 Tail Spike Protein, Gpv
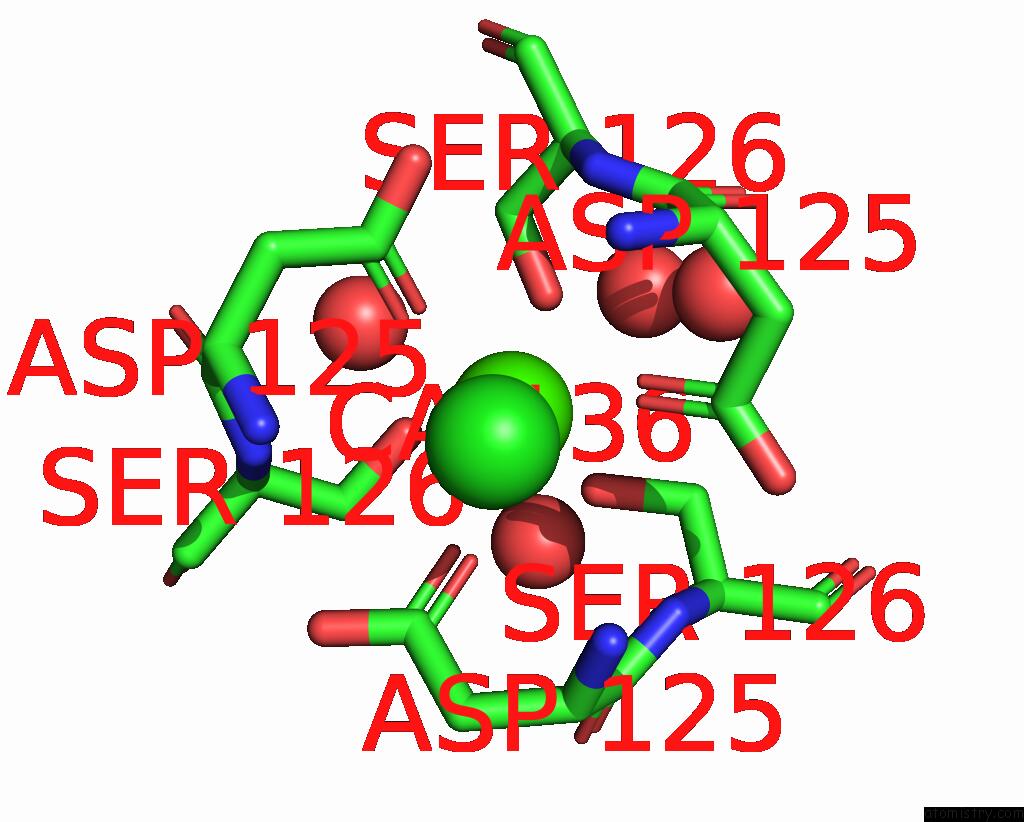
Mono view
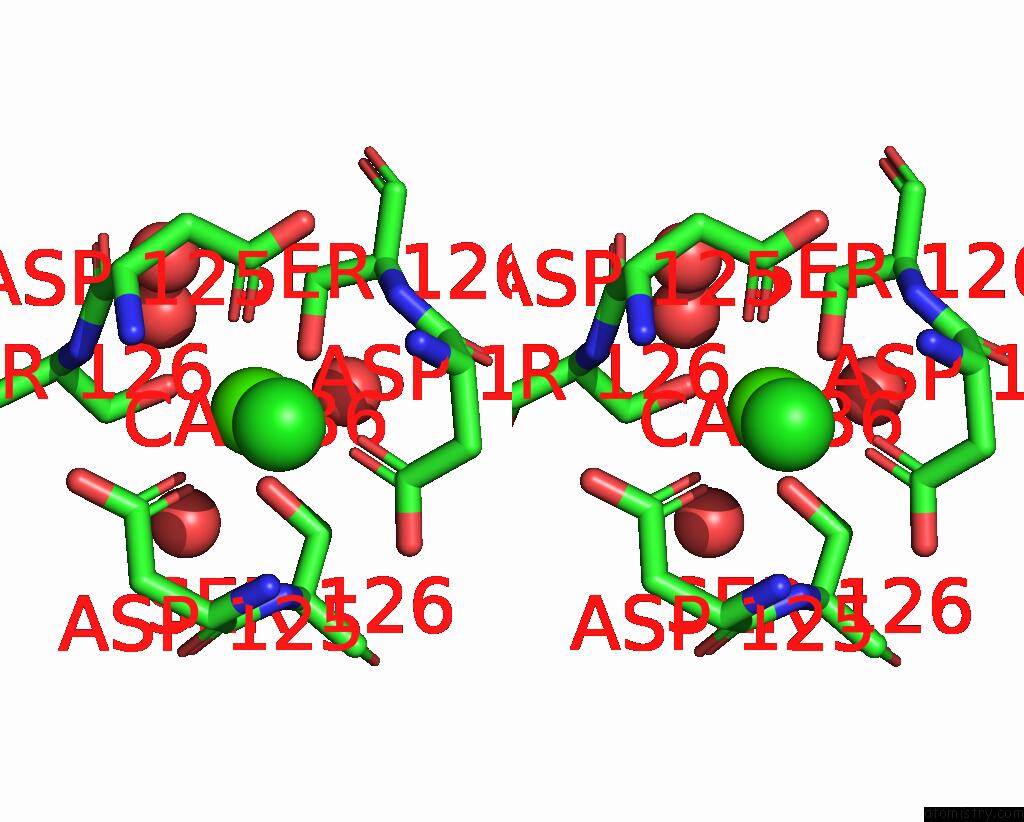
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Crystal Structure of A C-Terminal Domain of the Bacteriophage P2 Tail Spike Protein, Gpv within 5.0Å range:
|
Reference:
E.Yamashita,
A.Nakagawa,
J.Takahashi,
K.Tsunoda,
S.Yamada,
S.Takeda.
The Host-Binding Domain of the P2 Phage Tail Spike Reveals A Trimeric Iron-Binding Structure Acta Crystallogr.,Sect.F V. 67 837 2011.
ISSN: ESSN 1744-3091
PubMed: 21821878
DOI: 10.1107/S1744309111005999
Page generated: Tue Jul 8 10:54:07 2025
ISSN: ESSN 1744-3091
PubMed: 21821878
DOI: 10.1107/S1744309111005999
Last articles
Ca in 3GZTCa in 3HCM
Ca in 3HCI
Ca in 3HBV
Ca in 3HBZ
Ca in 3HBU
Ca in 3HB2
Ca in 3HBT
Ca in 3HB3
Ca in 3HAJ