Calcium »
PDB 6fzw-6guy »
6gp2 »
Calcium in PDB 6gp2: Ribonucleotide Reductase Class Ie R2 From Mesoplasma Florum, Dopa- Active Form
Protein crystallography data
The structure of Ribonucleotide Reductase Class Ie R2 From Mesoplasma Florum, Dopa- Active Form, PDB code: 6gp2
was solved by
V.Srinivas,
H.Lebrette,
D.Lundin,
Y.Kutin,
M.Sahlin,
M.Lerche,
J.Enrich,
R.M.M.Branca,
N.Cox,
B.M.Sjoberg,
M.Hogbom,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 42.73 / 1.48 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 176.162, 53.744, 79.284, 90.00, 108.58, 90.00 |
| R / Rfree (%) | 15.4 / 18.5 |
Calcium Binding Sites:
The binding sites of Calcium atom in the Ribonucleotide Reductase Class Ie R2 From Mesoplasma Florum, Dopa- Active Form
(pdb code 6gp2). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 2 binding sites of Calcium where determined in the Ribonucleotide Reductase Class Ie R2 From Mesoplasma Florum, Dopa- Active Form, PDB code: 6gp2:
Jump to Calcium binding site number: 1; 2;
In total 2 binding sites of Calcium where determined in the Ribonucleotide Reductase Class Ie R2 From Mesoplasma Florum, Dopa- Active Form, PDB code: 6gp2:
Jump to Calcium binding site number: 1; 2;
Calcium binding site 1 out of 2 in 6gp2
Go back to
Calcium binding site 1 out
of 2 in the Ribonucleotide Reductase Class Ie R2 From Mesoplasma Florum, Dopa- Active Form
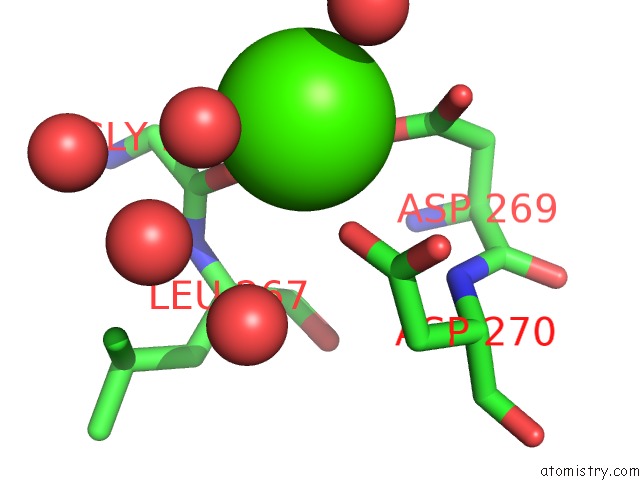
Mono view
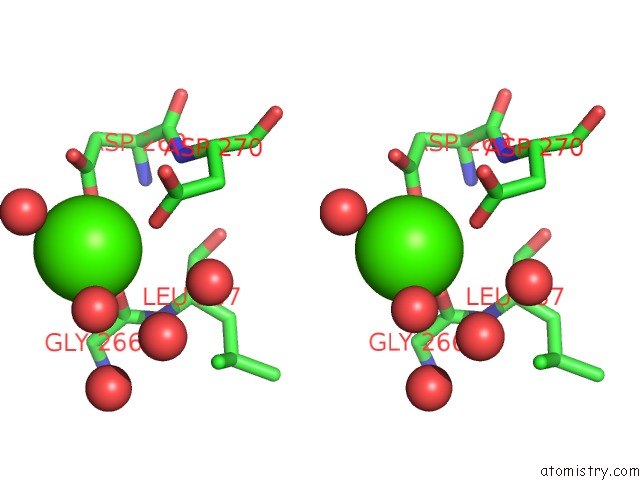
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Ribonucleotide Reductase Class Ie R2 From Mesoplasma Florum, Dopa- Active Form within 5.0Å range:
|
Calcium binding site 2 out of 2 in 6gp2
Go back to
Calcium binding site 2 out
of 2 in the Ribonucleotide Reductase Class Ie R2 From Mesoplasma Florum, Dopa- Active Form
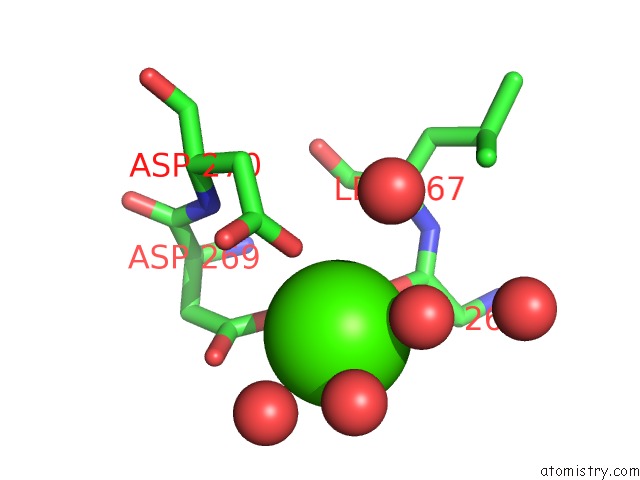
Mono view
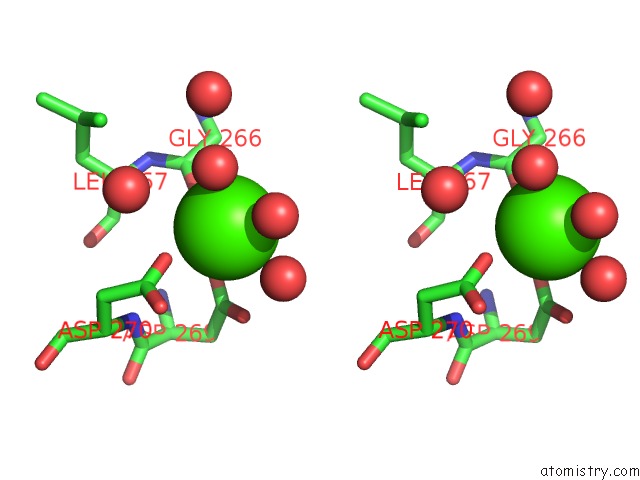
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Ribonucleotide Reductase Class Ie R2 From Mesoplasma Florum, Dopa- Active Form within 5.0Å range:
|
Reference:
V.Srinivas,
H.Lebrette,
D.Lundin,
Y.Kutin,
M.Sahlin,
M.Lerche,
J.Eirich,
R.M.M.Branca,
N.Cox,
B.M.Sjoberg,
M.Hogbom.
Metal-Free Ribonucleotide Reduction Powered By A Dopa Radical in Mycoplasma Pathogens. Nature V. 563 416 2018.
ISSN: ESSN 1476-4687
PubMed: 30429545
DOI: 10.1038/S41586-018-0653-6
Page generated: Wed Jul 9 14:25:41 2025
ISSN: ESSN 1476-4687
PubMed: 30429545
DOI: 10.1038/S41586-018-0653-6
Last articles
Ca in 7K4OCa in 7K4C
Ca in 7K4D
Ca in 7K4F
Ca in 7K44
Ca in 7K4B
Ca in 7K4A
Ca in 7JWX
Ca in 7K04
Ca in 7JYW