Calcium »
PDB 6q7o-6qps »
6qcl »
Calcium in PDB 6qcl: Citryl-Coa Lyase Core Module of Chlorobium Limicola Atp Citrate Lyase in Complex with Acetyl-Coa and L-Malate
Protein crystallography data
The structure of Citryl-Coa Lyase Core Module of Chlorobium Limicola Atp Citrate Lyase in Complex with Acetyl-Coa and L-Malate, PDB code: 6qcl
was solved by
K.Verstraete,
K.Verschueren,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 45.62 / 1.60 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 102.730, 100.817, 105.012, 90.00, 102.99, 90.00 |
| R / Rfree (%) | 15.7 / 17.7 |
Calcium Binding Sites:
The binding sites of Calcium atom in the Citryl-Coa Lyase Core Module of Chlorobium Limicola Atp Citrate Lyase in Complex with Acetyl-Coa and L-Malate
(pdb code 6qcl). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 2 binding sites of Calcium where determined in the Citryl-Coa Lyase Core Module of Chlorobium Limicola Atp Citrate Lyase in Complex with Acetyl-Coa and L-Malate, PDB code: 6qcl:
Jump to Calcium binding site number: 1; 2;
In total 2 binding sites of Calcium where determined in the Citryl-Coa Lyase Core Module of Chlorobium Limicola Atp Citrate Lyase in Complex with Acetyl-Coa and L-Malate, PDB code: 6qcl:
Jump to Calcium binding site number: 1; 2;
Calcium binding site 1 out of 2 in 6qcl
Go back to
Calcium binding site 1 out
of 2 in the Citryl-Coa Lyase Core Module of Chlorobium Limicola Atp Citrate Lyase in Complex with Acetyl-Coa and L-Malate
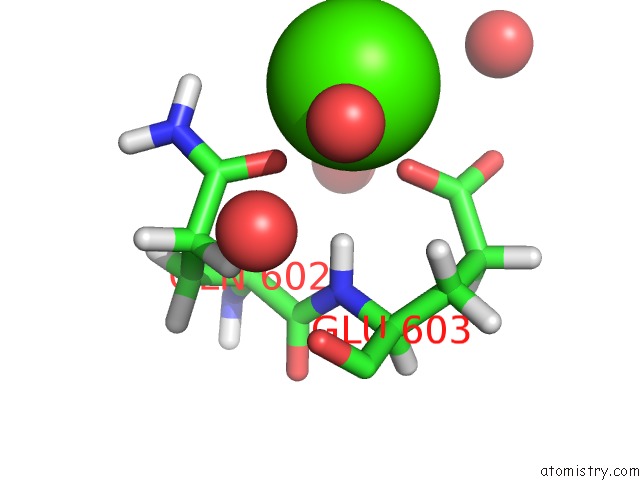
Mono view
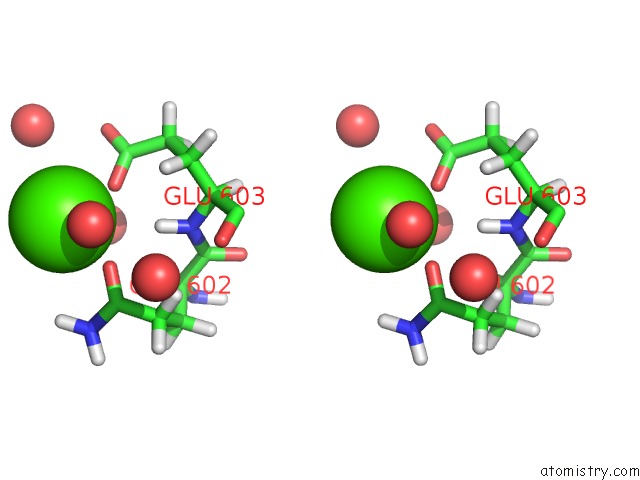
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Citryl-Coa Lyase Core Module of Chlorobium Limicola Atp Citrate Lyase in Complex with Acetyl-Coa and L-Malate within 5.0Å range:
|
Calcium binding site 2 out of 2 in 6qcl
Go back to
Calcium binding site 2 out
of 2 in the Citryl-Coa Lyase Core Module of Chlorobium Limicola Atp Citrate Lyase in Complex with Acetyl-Coa and L-Malate
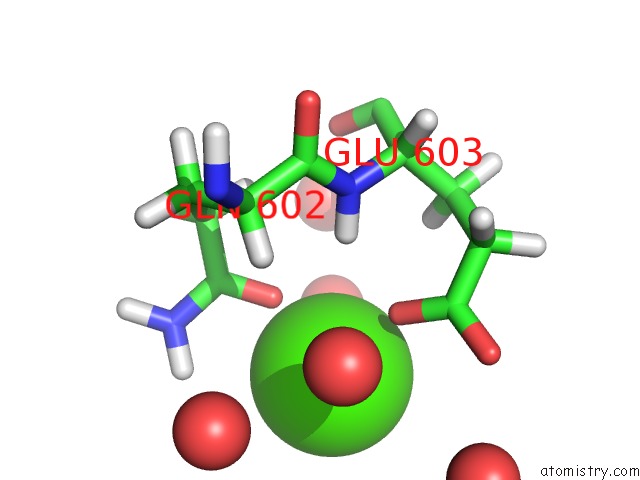
Mono view
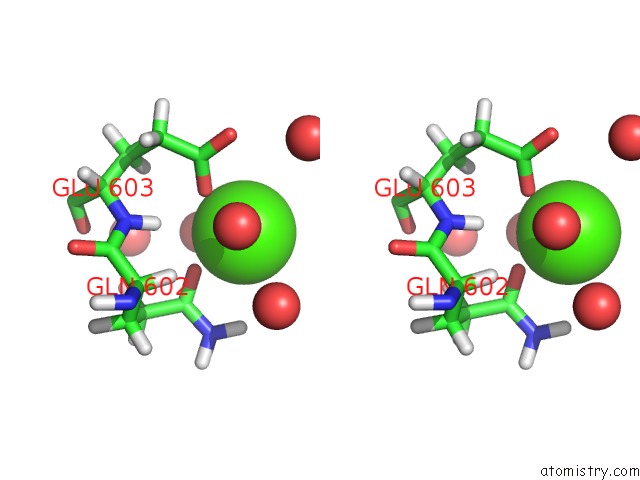
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Citryl-Coa Lyase Core Module of Chlorobium Limicola Atp Citrate Lyase in Complex with Acetyl-Coa and L-Malate within 5.0Å range:
|
Reference:
K.H.G.Verschueren,
C.Blanchet,
J.Felix,
A.Dansercoer,
D.De Vos,
Y.Bloch,
J.Van Beeumen,
D.Svergun,
I.Gutsche,
S.N.Savvides,
K.Verstraete.
Structure of Atp Citrate Lyase and the Origin of Citrate Synthase in the Krebs Cycle. Nature V. 568 571 2019.
ISSN: ESSN 1476-4687
PubMed: 30944476
DOI: 10.1038/S41586-019-1095-5
Page generated: Wed Jul 9 17:09:18 2025
ISSN: ESSN 1476-4687
PubMed: 30944476
DOI: 10.1038/S41586-019-1095-5
Last articles
Cl in 5L39Cl in 5L38
Cl in 5L35
Cl in 5L2U
Cl in 5L2N
Cl in 5L1E
Cl in 5L2K
Cl in 5L0K
Cl in 5L2M
Cl in 5L2J