Calcium »
PDB 6q7o-6qps »
6qpc »
Calcium in PDB 6qpc: Cryo-Em Structure of Calcium-Bound MTMEM16F Lipid Scramblase in Nanodisc
Calcium Binding Sites:
The binding sites of Calcium atom in the Cryo-Em Structure of Calcium-Bound MTMEM16F Lipid Scramblase in Nanodisc
(pdb code 6qpc). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 4 binding sites of Calcium where determined in the Cryo-Em Structure of Calcium-Bound MTMEM16F Lipid Scramblase in Nanodisc, PDB code: 6qpc:
Jump to Calcium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Calcium where determined in the Cryo-Em Structure of Calcium-Bound MTMEM16F Lipid Scramblase in Nanodisc, PDB code: 6qpc:
Jump to Calcium binding site number: 1; 2; 3; 4;
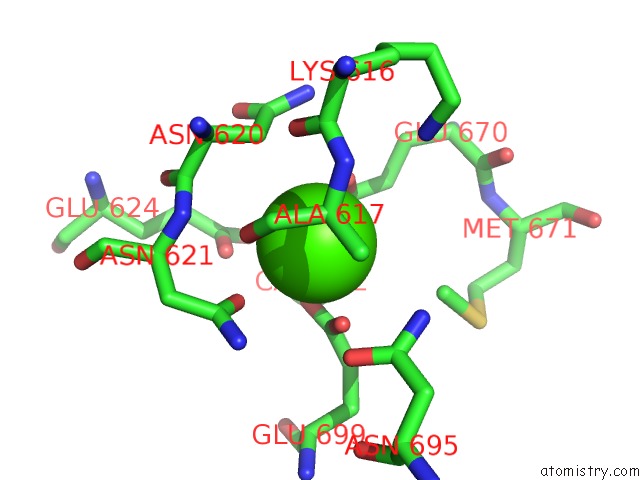
Calcium binding site 1 out of 4 in 6qpc
Go back to
Calcium binding site 1 out
of 4 in the Cryo-Em Structure of Calcium-Bound MTMEM16F Lipid Scramblase in Nanodisc
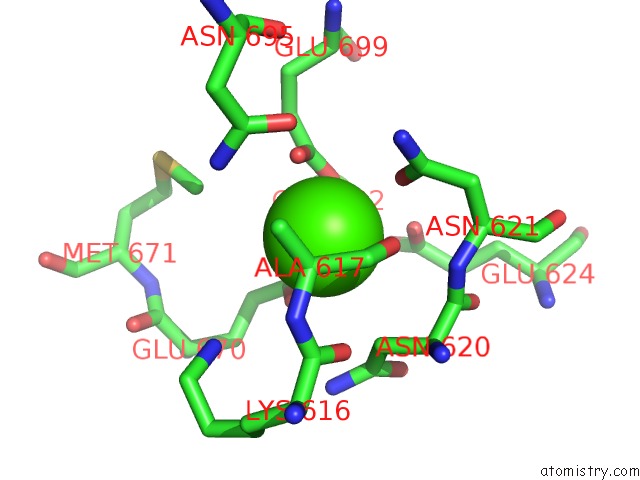
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Cryo-Em Structure of Calcium-Bound MTMEM16F Lipid Scramblase in Nanodisc within 5.0Å range:
|
Calcium binding site 2 out of 4 in 6qpc
Go back to
Calcium binding site 2 out
of 4 in the Cryo-Em Structure of Calcium-Bound MTMEM16F Lipid Scramblase in Nanodisc

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Cryo-Em Structure of Calcium-Bound MTMEM16F Lipid Scramblase in Nanodisc within 5.0Å range:
|
Calcium binding site 3 out of 4 in 6qpc
Go back to
Calcium binding site 3 out
of 4 in the Cryo-Em Structure of Calcium-Bound MTMEM16F Lipid Scramblase in Nanodisc
Mono view
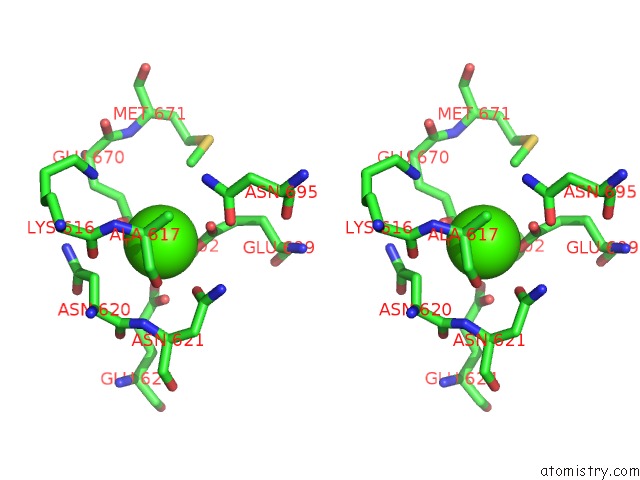
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 3 of Cryo-Em Structure of Calcium-Bound MTMEM16F Lipid Scramblase in Nanodisc within 5.0Å range:
|
Calcium binding site 4 out of 4 in 6qpc
Go back to
Calcium binding site 4 out
of 4 in the Cryo-Em Structure of Calcium-Bound MTMEM16F Lipid Scramblase in Nanodisc

Mono view
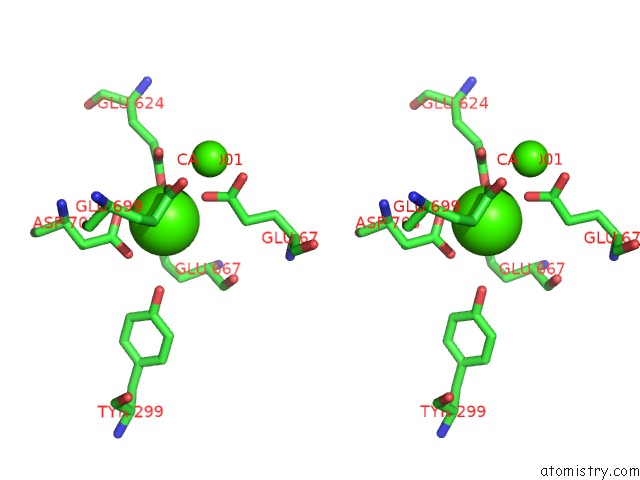
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 4 of Cryo-Em Structure of Calcium-Bound MTMEM16F Lipid Scramblase in Nanodisc within 5.0Å range:
|
Reference:
C.Alvadia,
N.K.Lim,
V.Clerico Mosina,
G.T.Oostergetel,
R.Dutzler,
C.Paulino.
Cryo-Em Structures and Functional Characterization of the Murine Lipid Scramblase TMEM16F. Elife V. 8 2019.
ISSN: ESSN 2050-084X
PubMed: 30785399
DOI: 10.7554/ELIFE.44365
Page generated: Wed Jul 9 17:16:22 2025
ISSN: ESSN 2050-084X
PubMed: 30785399
DOI: 10.7554/ELIFE.44365
Last articles
Ca in 7LN7Ca in 7LJB
Ca in 7LJJ
Ca in 7LK7
Ca in 7LCE
Ca in 7LJA
Ca in 7LJ5
Ca in 7LJ4
Ca in 7LJ2
Ca in 7LHA