Calcium »
PDB 8ica-8iww »
8ihy »
Calcium in PDB 8ihy: X-Ray Crystal Structure of Q387E Mutant of Endo-1,4-Beta Glucanase From Eisenia Fetida
Protein crystallography data
The structure of X-Ray Crystal Structure of Q387E Mutant of Endo-1,4-Beta Glucanase From Eisenia Fetida, PDB code: 8ihy
was solved by
C.Kuroki,
Y.Hirano,
M.Nakazawa,
T.Sakamoto,
T.Tamada,
M.Ueda,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 40.29 / 1.60 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 53.36, 71.639, 55.184, 90, 114.04, 90 |
| R / Rfree (%) | 14.7 / 17.7 |
Other elements in 8ihy:
The structure of X-Ray Crystal Structure of Q387E Mutant of Endo-1,4-Beta Glucanase From Eisenia Fetida also contains other interesting chemical elements:
| Magnesium | (Mg) | 1 atom |
| Sodium | (Na) | 1 atom |
Calcium Binding Sites:
The binding sites of Calcium atom in the X-Ray Crystal Structure of Q387E Mutant of Endo-1,4-Beta Glucanase From Eisenia Fetida
(pdb code 8ihy). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total only one binding site of Calcium was determined in the X-Ray Crystal Structure of Q387E Mutant of Endo-1,4-Beta Glucanase From Eisenia Fetida, PDB code: 8ihy:
In total only one binding site of Calcium was determined in the X-Ray Crystal Structure of Q387E Mutant of Endo-1,4-Beta Glucanase From Eisenia Fetida, PDB code: 8ihy:
Calcium binding site 1 out of 1 in 8ihy
Go back to
Calcium binding site 1 out
of 1 in the X-Ray Crystal Structure of Q387E Mutant of Endo-1,4-Beta Glucanase From Eisenia Fetida
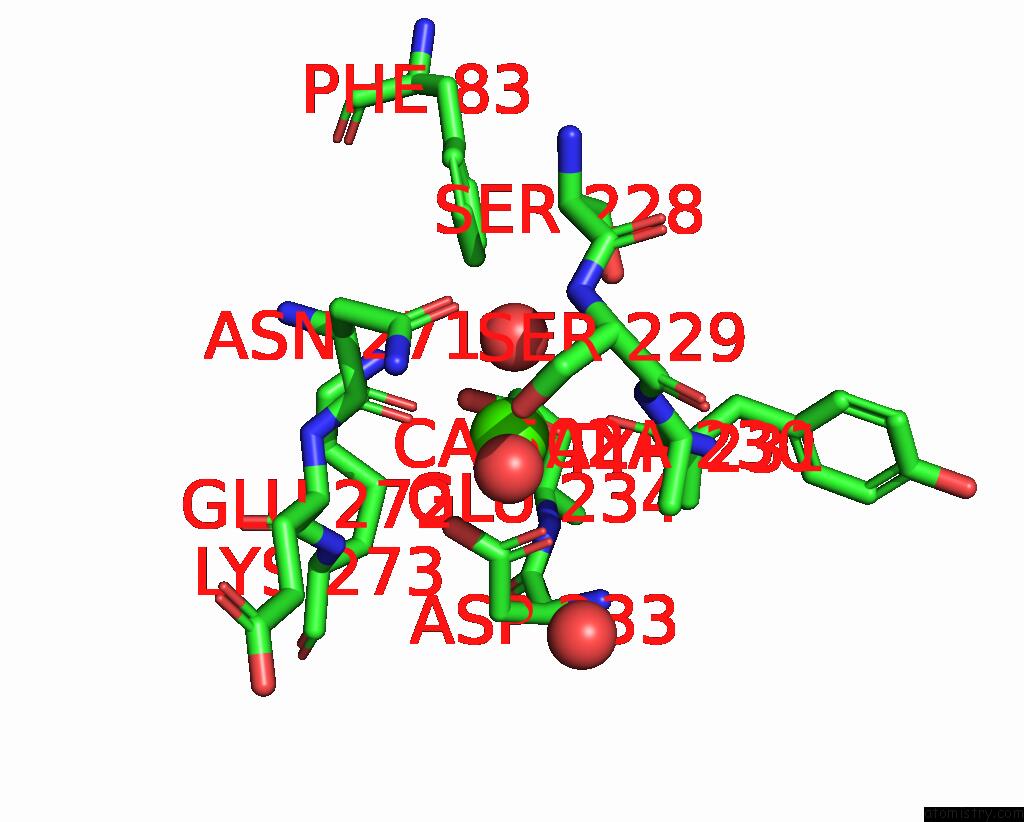
Mono view
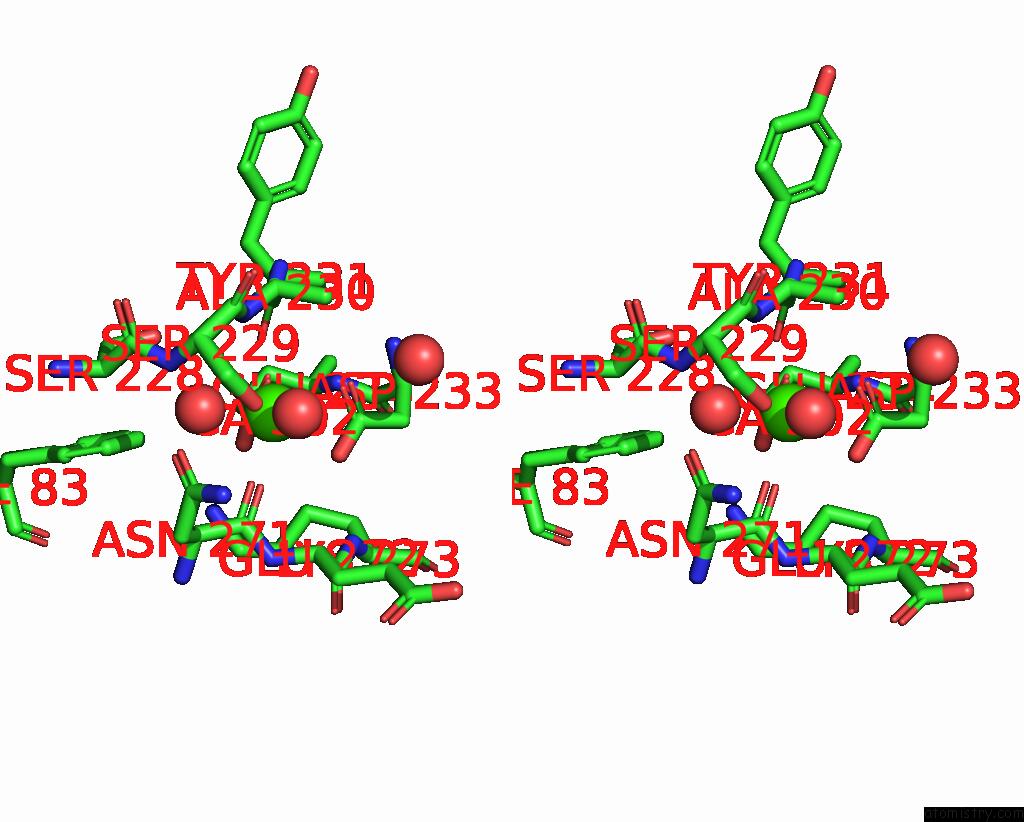
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of X-Ray Crystal Structure of Q387E Mutant of Endo-1,4-Beta Glucanase From Eisenia Fetida within 5.0Å range:
|
Reference:
C.Kuroki,
Y.Hirano,
M.Nakazawa,
T.Sakamoto,
T.Tamada,
M.Ueda.
A Single Mutation ASP43ARG Was Increased 2.5-Fold the Catalytic Activity and Maintained the Stability of Cold-Adapted Endo-1,4-Beta Glucanase (Ef-EG2) From Eisenia Fetida. Curr Res Biotechnol V. 5 2023.
DOI: 10.1016/J.CRBIOT.2023.100126
Page generated: Thu Jul 10 05:13:58 2025
DOI: 10.1016/J.CRBIOT.2023.100126
Last articles
Cl in 5IMECl in 5ILF
Cl in 5ILR
Cl in 5ILM
Cl in 5ILC
Cl in 5IKQ
Cl in 5IKT
Cl in 5IKJ
Cl in 5IKF
Cl in 5IKM