Calcium »
PDB 2yi1-2z8s »
2z4e »
Calcium in PDB 2z4e: Crystal Structure of D-Dimer From Human Fibrin Complexed with Gly-His- Arg-Pro-Tyr-Amide
Protein crystallography data
The structure of Crystal Structure of D-Dimer From Human Fibrin Complexed with Gly-His- Arg-Pro-Tyr-Amide, PDB code: 2z4e
was solved by
R.F.Doolittle,
L.Pandi,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 30.00 / 2.70 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 54.630, 148.690, 232.680, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 21.6 / 27.5 |
Calcium Binding Sites:
The binding sites of Calcium atom in the Crystal Structure of D-Dimer From Human Fibrin Complexed with Gly-His- Arg-Pro-Tyr-Amide
(pdb code 2z4e). This binding sites where shown within
5.0 Angstroms radius around Calcium atom.
In total 8 binding sites of Calcium where determined in the Crystal Structure of D-Dimer From Human Fibrin Complexed with Gly-His- Arg-Pro-Tyr-Amide, PDB code: 2z4e:
Jump to Calcium binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
In total 8 binding sites of Calcium where determined in the Crystal Structure of D-Dimer From Human Fibrin Complexed with Gly-His- Arg-Pro-Tyr-Amide, PDB code: 2z4e:
Jump to Calcium binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
Calcium binding site 1 out of 8 in 2z4e
Go back to
Calcium binding site 1 out
of 8 in the Crystal Structure of D-Dimer From Human Fibrin Complexed with Gly-His- Arg-Pro-Tyr-Amide
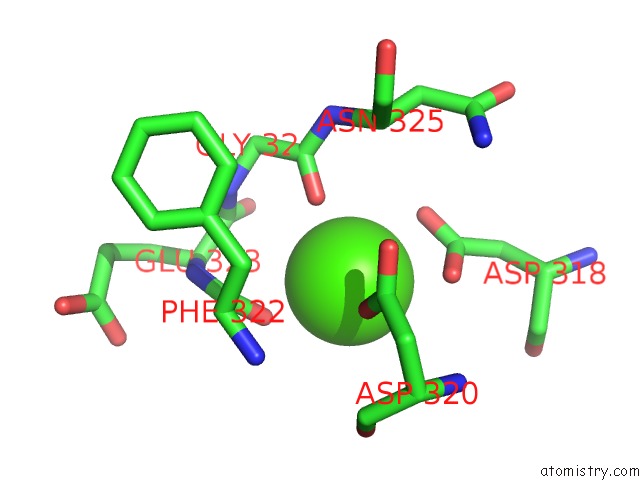
Mono view
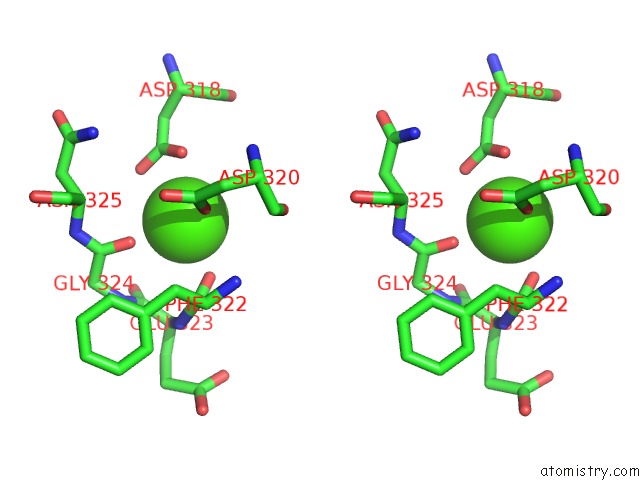
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 1 of Crystal Structure of D-Dimer From Human Fibrin Complexed with Gly-His- Arg-Pro-Tyr-Amide within 5.0Å range:
|
Calcium binding site 2 out of 8 in 2z4e
Go back to
Calcium binding site 2 out
of 8 in the Crystal Structure of D-Dimer From Human Fibrin Complexed with Gly-His- Arg-Pro-Tyr-Amide
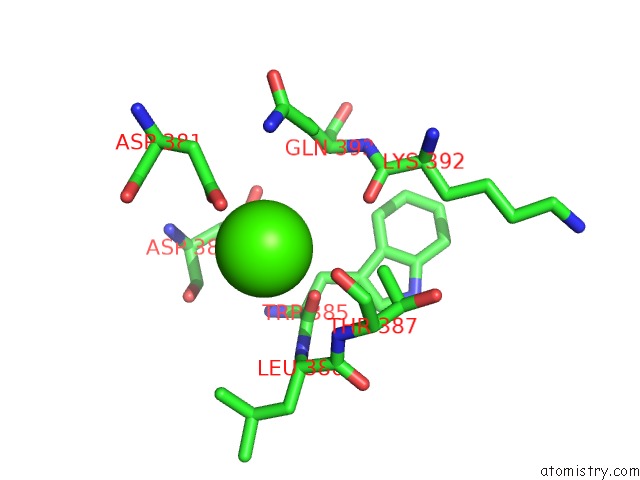
Mono view
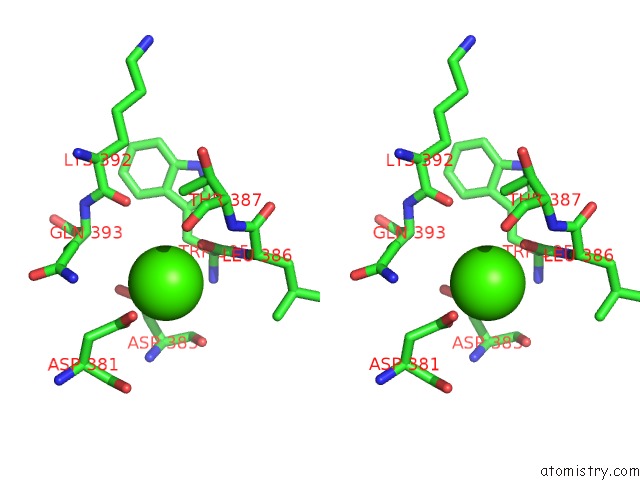
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 2 of Crystal Structure of D-Dimer From Human Fibrin Complexed with Gly-His- Arg-Pro-Tyr-Amide within 5.0Å range:
|
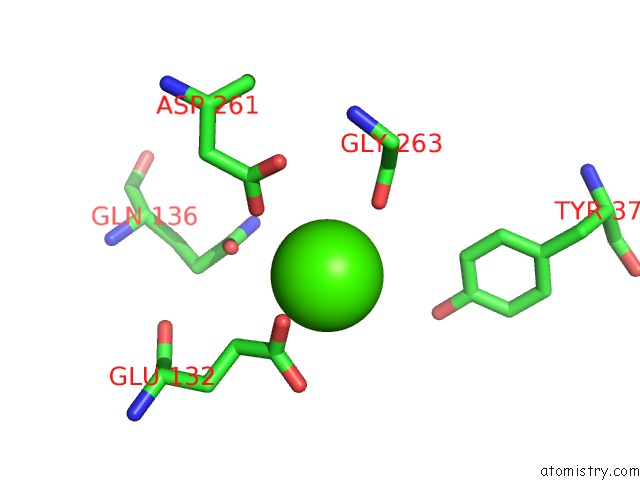
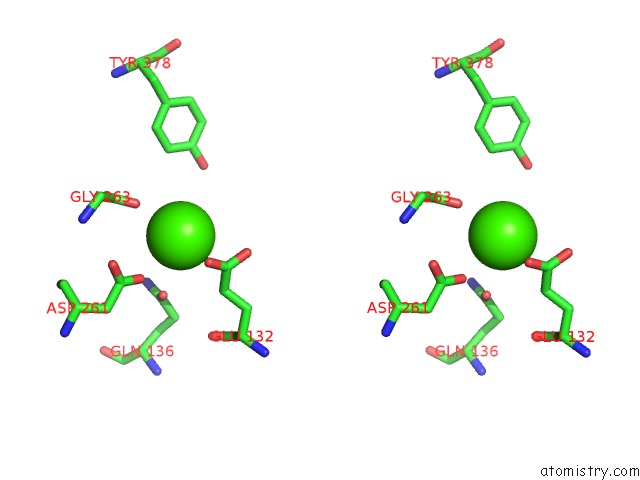
Calcium binding site 3 out of 8 in 2z4e
Go back to
Calcium binding site 3 out
of 8 in the Crystal Structure of D-Dimer From Human Fibrin Complexed with Gly-His- Arg-Pro-Tyr-Amide
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 3 of Crystal Structure of D-Dimer From Human Fibrin Complexed with Gly-His- Arg-Pro-Tyr-Amide within 5.0Å range:
|
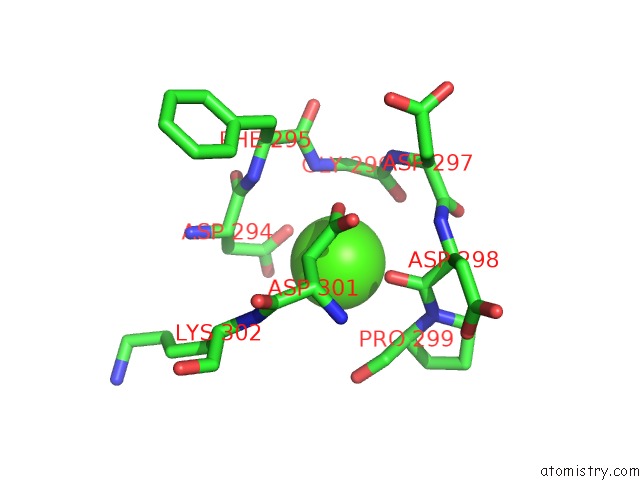
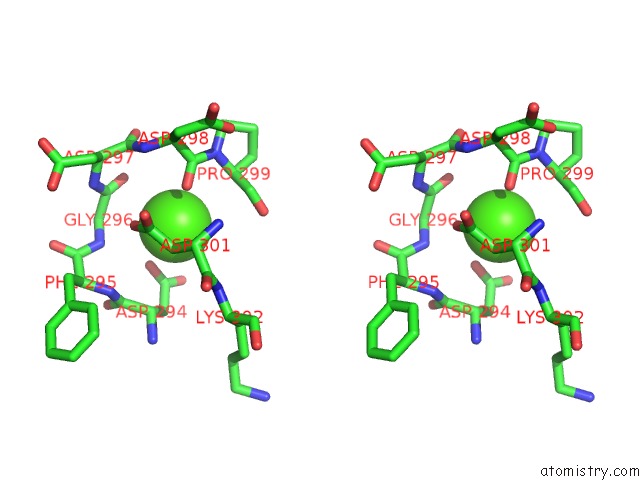
Calcium binding site 4 out of 8 in 2z4e
Go back to
Calcium binding site 4 out
of 8 in the Crystal Structure of D-Dimer From Human Fibrin Complexed with Gly-His- Arg-Pro-Tyr-Amide
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 4 of Crystal Structure of D-Dimer From Human Fibrin Complexed with Gly-His- Arg-Pro-Tyr-Amide within 5.0Å range:
|
Calcium binding site 5 out of 8 in 2z4e
Go back to
Calcium binding site 5 out
of 8 in the Crystal Structure of D-Dimer From Human Fibrin Complexed with Gly-His- Arg-Pro-Tyr-Amide
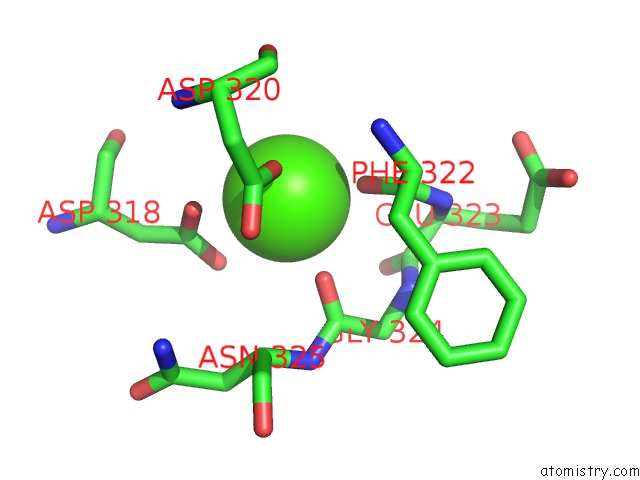
Mono view
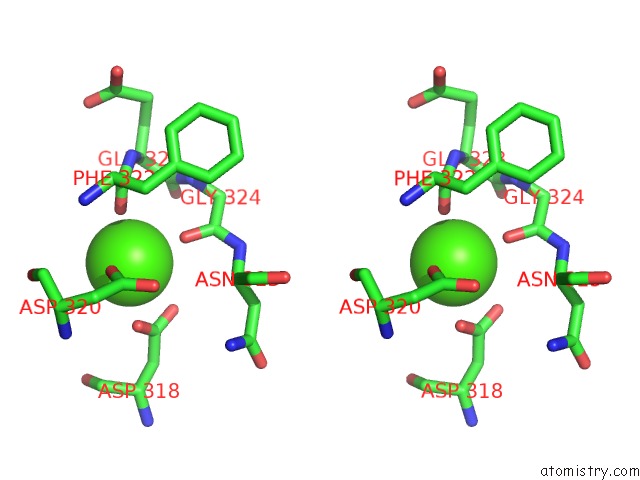
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 5 of Crystal Structure of D-Dimer From Human Fibrin Complexed with Gly-His- Arg-Pro-Tyr-Amide within 5.0Å range:
|
Calcium binding site 6 out of 8 in 2z4e
Go back to
Calcium binding site 6 out
of 8 in the Crystal Structure of D-Dimer From Human Fibrin Complexed with Gly-His- Arg-Pro-Tyr-Amide
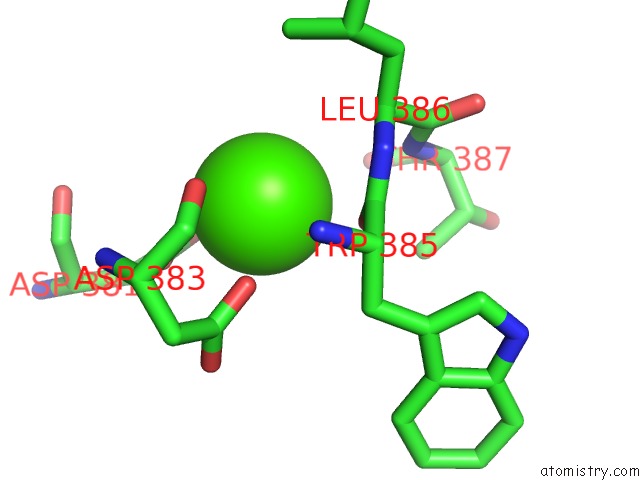
Mono view
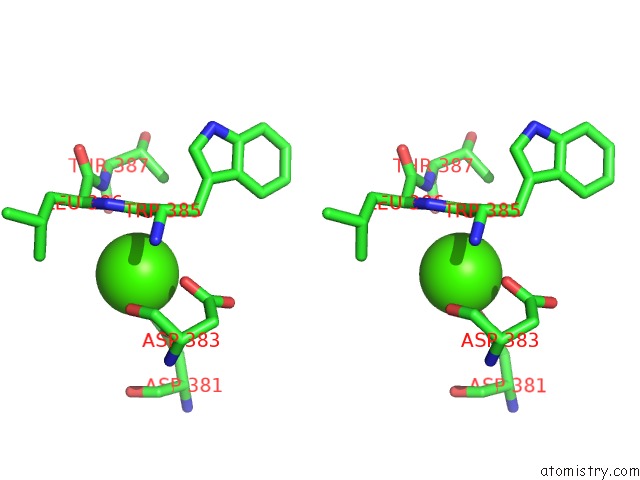
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 6 of Crystal Structure of D-Dimer From Human Fibrin Complexed with Gly-His- Arg-Pro-Tyr-Amide within 5.0Å range:
|
Calcium binding site 7 out of 8 in 2z4e
Go back to
Calcium binding site 7 out
of 8 in the Crystal Structure of D-Dimer From Human Fibrin Complexed with Gly-His- Arg-Pro-Tyr-Amide
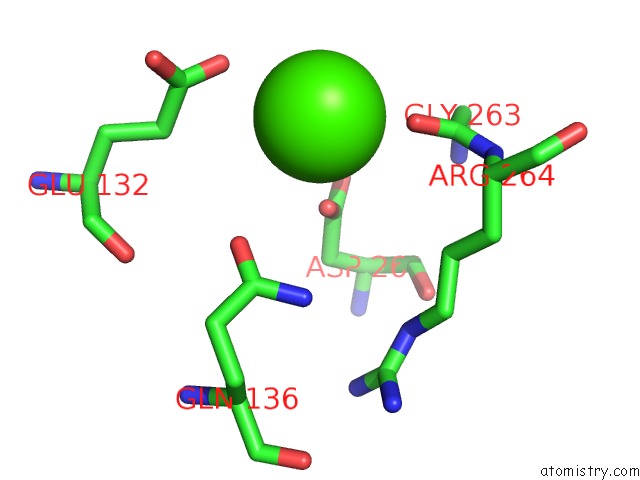
Mono view
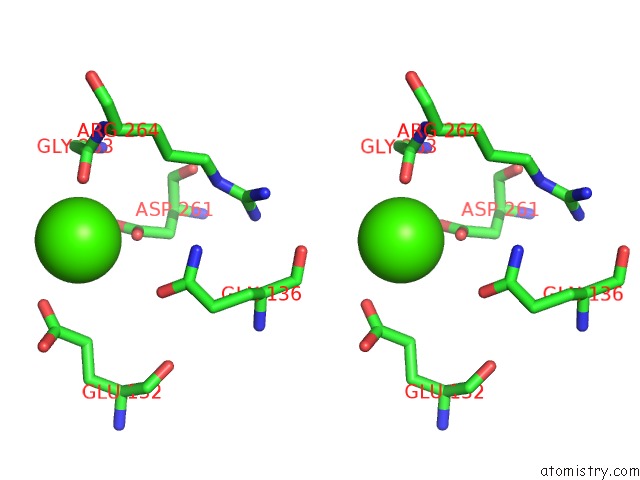
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 7 of Crystal Structure of D-Dimer From Human Fibrin Complexed with Gly-His- Arg-Pro-Tyr-Amide within 5.0Å range:
|
Calcium binding site 8 out of 8 in 2z4e
Go back to
Calcium binding site 8 out
of 8 in the Crystal Structure of D-Dimer From Human Fibrin Complexed with Gly-His- Arg-Pro-Tyr-Amide
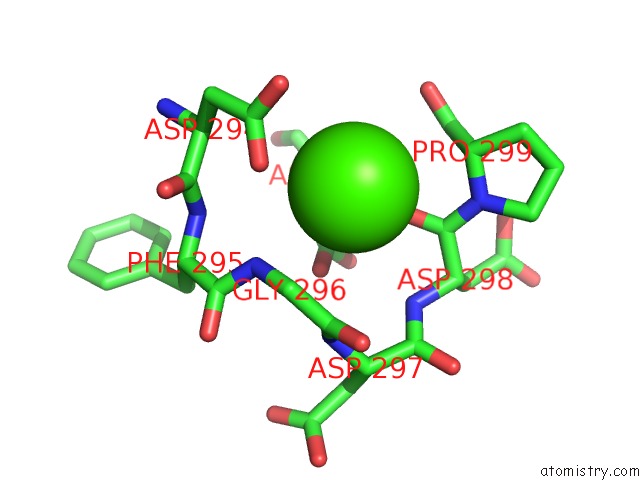
Mono view
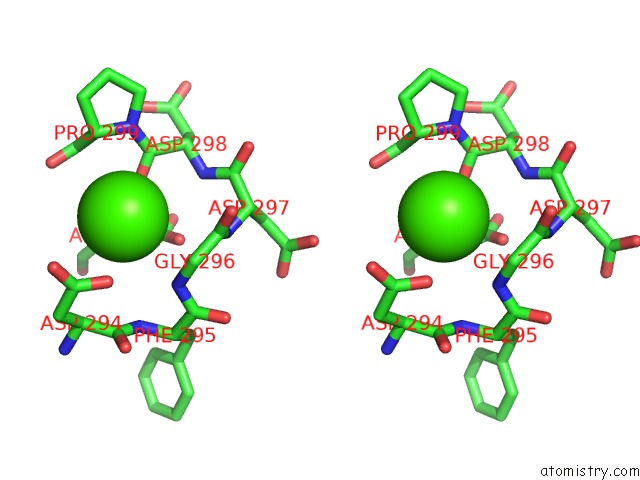
Stereo pair view

Mono view

Stereo pair view
A full contact list of Calcium with other atoms in the Ca binding
site number 8 of Crystal Structure of D-Dimer From Human Fibrin Complexed with Gly-His- Arg-Pro-Tyr-Amide within 5.0Å range:
|
Reference:
R.F.Doolittle,
L.Pandi.
Probing the Beta-Chain Hole of Fibrinogen with Synthetic Peptides That Differ at Their Amino Termini Biochemistry V. 46 10033 2007.
ISSN: ISSN 0006-2960
PubMed: 17688324
DOI: 10.1021/BI7010916
Page generated: Tue Jul 8 09:49:49 2025
ISSN: ISSN 0006-2960
PubMed: 17688324
DOI: 10.1021/BI7010916
Last articles
Ca in 3LJMCa in 3LJG
Ca in 3LJK
Ca in 3LJJ
Ca in 3LIR
Ca in 3LIW
Ca in 3LIP
Ca in 3LIJ
Ca in 3LIK
Ca in 3LIL